Cryo-electron microscopy structure of adenovirus type 2 temperature-sensitive mutant 1 reveals insight into the cell entry defect
- PMID: 19458007
- PMCID: PMC2708647
- DOI: 10.1128/JVI.00331-09
Cryo-electron microscopy structure of adenovirus type 2 temperature-sensitive mutant 1 reveals insight into the cell entry defect
Abstract
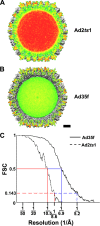
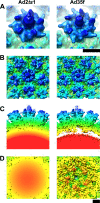
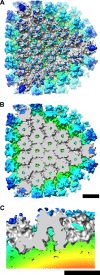
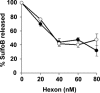
The structure of the adenovirus type 2 temperature-sensitive mutant 1 (Ad2ts1) was determined to a resolution of 10 A by cryo-electron microscopy single-particle reconstruction. Ad2ts1 was prepared at a nonpermissive temperature and contains the precursor forms of the capsid proteins IIIa, VI, and VIII; the core proteins VII, X (mu), and terminal protein (TP); and the L1-52K protein. Cell entry studies have shown that although Ad2ts1 can bind the coxsackievirus and Ad receptor and undergo internalization via alphav integrins, this mutant does not escape from the early endosome and is targeted for degradation. Comparison of the Ad2ts1 structure to that of mature Ad indicates that Ad2ts1 has a different core architecture. The Ad2ts1 core is closely associated with the icosahedral capsid, a connection which may be mediated by preproteins IIIa and VI. Density within hexon cavities is assigned to preprotein VI, and membrane disruption assays show that hexon shields the lytic activity of both the mature and precursor forms of protein VI. The internal surface of the penton base in Ad2ts1 appears to be anchored to the core by interactions with preprotein IIIa. Our structural analyses suggest that these connections to the core inhibit the release of the vertex proteins and lead to the cell entry defect of Ad2ts1.
Figures



References
-
- Adiga, U., W. T. Baxter, R. J. Hall, B. Rockel, B. K. Rath, J. Frank, and R. Glaeser. 2005. Particle picking by segmentation: a comparative study with SPIDER-based manual particle picking. J. Struct. Biol. 152211-220. - PubMed
-
- Anderson, C. W. 1990. The proteinase polypeptide of adenovirus serotype 2 virions. Virology 177259-272. - PubMed
-
- Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 2751320-1323. - PubMed
-
- Brown, M. T., W. J. McGrath, D. L. Toledo, and W. F. Mangel. 1996. Different modes of inhibition of human adenovirus proteinase, probably a cysteine proteinase, by bovine pancreatic trypsin inhibitor. FEBS Lett. 388233-237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

