Conserved introns reveal novel transcripts in Drosophila melanogaster
- PMID: 19458021
- PMCID: PMC2704441
- DOI: 10.1101/gr.090050.108
Conserved introns reveal novel transcripts in Drosophila melanogaster
Abstract
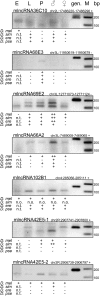
Noncoding RNAs that are-like mRNAs-spliced, capped, and polyadenylated have important functions in cellular processes. The inventory of these mRNA-like noncoding RNAs (mlncRNAs), however, is incomplete even in well-studied organisms, and so far, no computational methods exist to predict such RNAs from genomic sequences only. The subclass of these transcripts that is evolutionarily conserved usually has conserved intron positions. We demonstrate here that a genome-wide comparative genomics approach searching for short conserved introns is capable of identifying conserved transcripts with a high specificity. Our approach requires neither an open reading frame nor substantial sequence or secondary structure conservation in the surrounding exons. Thus it identifies spliced transcripts in an unbiased way. After applying our approach to insect genomes, we predict 369 introns outside annotated coding transcripts, of which 131 are confirmed by expressed sequence tags (ESTs) and/or noncoding FlyBase transcripts. Of the remaining 238 novel introns, about half are associated with protein-coding genes-either extending coding or untranslated regions or likely belonging to unannotated coding genes. The remaining 129 introns belong to novel mlncRNAs that are largely unstructured. Using RT-PCR, we verified seven of 12 tested introns in novel mlncRNAs and 11 of 17 introns in novel coding genes. The expression level of all verified mlncRNA transcripts is low but varies during development, which suggests regulation. As conserved introns indicate both purifying selection on the exon-intron structure and conserved expression of the transcript in related species, the novel mlncRNAs are good candidates for functional transcripts.
Figures





References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. - PubMed
-
- Arya R, Mallik M, Lakhotia SC. Heat shock genes: Integrating cell survival and death. J Biosci. 2007;32:595–610. - PubMed
-
- Badger JH, Olsen GJ. Critica: Coding region identification tool invoking comparative analysis. Mol Biol Evol. 1999;16:512–524. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
