Type III protein secretion in plant pathogenic bacteria
- PMID: 19458111
- PMCID: PMC2719110
- DOI: 10.1104/pp.109.139089
Type III protein secretion in plant pathogenic bacteria
Figures
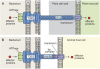

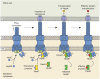
References
-
- Abrahams JP, Leslie AG, Lutter R, Walker JE (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370 621–628 - PubMed
-
- Akeda Y, Galán JE (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437 911–915 - PubMed
-
- Alfano JR, Collmer A (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42 385–414 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

