Data perturbation independent diagnosis and validation of breast cancer subtypes using clustering and patterns
- PMID: 19458770
- PMCID: PMC2675483
Data perturbation independent diagnosis and validation of breast cancer subtypes using clustering and patterns
Abstract
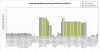
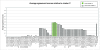
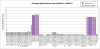
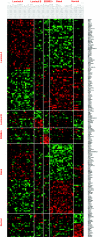
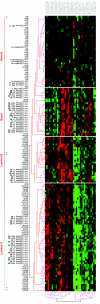
Molecular stratification of disease based on expression levels of sets of genes can help guide therapeutic decisions if such classifications can be shown to be stable against variations in sample source and data perturbation. Classifications inferred from one set of samples in one lab should be able to consistently stratify a different set of samples in another lab. We present a method for assessing such stability and apply it to the breast cancer (BCA) datasets of Sorlie et al. 2003 and Ma et al. 2003. We find that within the now commonly accepted BCA categories identified by Sorlie et al. Luminal A and Basal are robust, but Luminal B and ERBB2+ are not. In particular, 36% of the samples identified as Luminal B and 55% identified as ERBB2+ cannot be assigned an accurate category because the classification is sensitive to data perturbation. We identify a "core cluster" of samples for each category, and from these we determine "patterns" of gene expression that distinguish the core clusters from each other. We find that the best markers for Luminal A and Basal are (ESR1, LIV1, GATA-3) and (CCNE1, LAD1, KRT5), respectively. Pathways enriched in the patterns regulate apoptosis, tissue remodeling and the immune response. We use a different dataset (Ma et al. 2003) to test the accuracy with which samples can be allocated to the four disease subtypes. We find, as expected, that the classification of samples identified as Luminal A and Basal is robust but classification into the other two subtypes is not.
Keywords: Breast cancer; Clusters; Diagnosis; Multi-gene Biomarkers; Patterns.
Figures




References
References for Supplementary Information III
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B. 1995;57:289–300.
-
- Bonferroni CE. In Studi in Onore del Professore Salvatore Ortu Carboni; Rome: Italy: 1935. Il calcolo delle assicurazioni su gruppi di teste; pp. 13–60.
-
- Dudoit S, Popper Shaffer J, Boldrick JC. Multiple hypothesis testing in microarray experiments. Statistical Science. 2003;18:71–103.
References for Supplementary Information II
-
- Pavilidis P, Qin J, Arango V, et al. Using the gene ontology for microarray data mining: A comparison of methods and application to age effects in human prefrontal cortex. Neurochem. Res. 2004;29:1213–22. - PubMed
References
-
- Abd El-Rehim DM, Ball G, Pinder SE, et al. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int. J.Cancer. 2005;116:340–50. - PubMed
-
- Ahnstrom M, Nordenskjold B, Rutqvist LE, et al. Role of cyclin D1 in ErbB2-positive breast cancer and tamoxifen resistance. Breast Cancer Res. Treat. 2005;91:145–51. - PubMed
-
- Alexe G, Alexe S, Crama Y, et al. Consensus algorithms for the generation of all maximal bicliques. Disc. Appl. Math. 2004;145:11–21.
-
- Alexe G, Hammer PL. Spanned patterns in logical analysis of data. Discr. Appl. Math. 2005;154:1039–49.
-
- Alexe G, Bhanot G, Venkataraghavan B, et al. A robust meta-classification strategy for cancer diagnosis from gene expression data. Proc IEEE Comput Syst Bioinform Conf. 2005a:322–5. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
