Functional significance of the hemadsorption activity of influenza virus neuraminidase and its alteration in pandemic viruses
- PMID: 19458903
- PMCID: PMC2691527
- DOI: 10.1007/s00705-009-0393-x
Functional significance of the hemadsorption activity of influenza virus neuraminidase and its alteration in pandemic viruses
Abstract
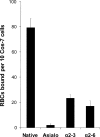
Human influenza viruses derive their genes from avian viruses. The neuraminidase (NA) of the avian viruses has, in addition to the catalytic site, a separate sialic acid binding site (hemadsorption site) that is not present in human viruses. The biological significance of the NA hemadsorption activity in avian influenza viruses remained elusive. A sequence database analysis revealed that the NAs of the majority of human H2N2 viruses isolated during the influenza pandemic of 1957 differ from their putative avian precursor by amino acid substitutions in the hemadsorption site. We found that the NA of a representative pandemic virus A/Singapore/1/57 (H2N2) lacks hemadsorption activity and that a single reversion to the avian-virus-like sequence (N367S) restores hemadsorption. Using this hemadsorption-positive NA, we generated three NA variants with substitutions S370L, N400S and W403R that have been found in the hemadsorption site of human H2N2 viruses. Each substitution abolished hemadsorption activity. Although, there was no correlation between hemadsorption activity of the NA variants and their enzymatic activity with respect to monovalent substrates, all four hemadsorption-negative NAs desialylated macromolecular substrates significantly slower than did the hemadsorption-positive counterpart. The NA of the 1918 pandemic virus A/Brevig Mission/1/18 (H1N1) also differed from avian N1 NAs by reduced hemadsorption activity and less efficient hydrolysis of macromolecular substrates. Our data indicate that the hemadsorption site serves to enhance the catalytic efficiency of NA and they suggest that, in addition to changes in the receptor-binding specificity of the hemagglutinin, alterations of the NA are needed for the emergence of pandemic influenza viruses.
Figures




References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/j.virol.2006.10.037', 'is_inner': False, 'url': 'https://doi.org/10.1016/j.virol.2006.10.037'}, {'type': 'PMC', 'value': 'PMC2735206', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC2735206/'}, {'type': 'PubMed', 'value': '17157891', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17157891/'}]}
- Aamir UB, Wernery U, Ilyushina N, Webster RG (2007) Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology 361:45–55 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1002/prot.340060402', 'is_inner': False, 'url': 'https://doi.org/10.1002/prot.340060402'}, {'type': 'PubMed', 'value': '2482974', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/2482974/'}]}
- Air GM, Laver WG (1989) The neuraminidase of influenza virus. Proteins 6:341–356 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1006/viro.1995.1401', 'is_inner': False, 'url': 'https://doi.org/10.1006/viro.1995.1401'}, {'type': 'PubMed', 'value': '7645221', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/7645221/'}]}
- Air GM, Laver WG (1995) Red cells bound to influenza virus N9 neuraminidase are not released by the N9 neuraminidase activity. Virology 211:278–284 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1002/prot.340020205', 'is_inner': False, 'url': 'https://doi.org/10.1002/prot.340020205'}, {'type': 'PubMed', 'value': '3447170', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/3447170/'}]}
- Baker AT, Varghese JN, Laver WG, Air GM, Colman PM (1987) Three-dimensional structure of neuraminidase of subtype N9 from an avian influenza virus. Proteins 2:111–117 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/0042-6822(91)90003-T', 'is_inner': False, 'url': 'https://doi.org/10.1016/0042-6822(91)90003-t'}, {'type': 'PubMed', 'value': '1984642', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/1984642/'}]}
- Baum LG, Paulson JC (1991) The N2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology 180:10–15 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

