NDUFS4: creation of a mouse model mimicking a Complex I disorder
- PMID: 19460290
- PMCID: PMC2783808
- DOI: 10.1016/j.mito.2009.02.001
NDUFS4: creation of a mouse model mimicking a Complex I disorder
Abstract
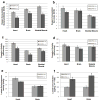
The Complex I NADH dehydrogenase-ubiquinone-FeS 4 (NDUFS4) subunit gene is involved in proper Complex I function such that the loss of NDUFS4 decreases Complex I activity resulting in mitochondrial disease. Therefore, a mouse model harboring a point mutation in the NDUFS4 gene was created. An embryonic lethal phenotype was observed in homozygous (NDUFS4(-/-)) mutant fetuses. Mitochondrial function was impaired in heterozygous animals based on oxygen consumption, and Complex I activity in NDUFS4 mouse mitochondria. Decreased Complex I activity with unaltered Complex II activity, along with an accumulation of lactate, were consistent with Complex I disorders in this mouse model.
Figures




References
-
- Agostino A, Invernizzi F, Tiveron C, Fagiolari G, Prelle A, Lamantea E, Giavazzi A, Battaglia G, Tatangelo L, Tiranti V, Zeviani M. Constitutive knockout of Surf1 is associated with high embryonic lethality, mitochondrial disease and cytochrome c oxidase deficiency in mice. Hum Mol Genet. 2003;12:399–413. - PubMed
-
- Aure K, Fayet G, Leroy JP, Lacene E, Romero NB, Lombes A. Apoptosis in mitochondrial myopathies is linked to mitochondrial proliferation. Brain. 2006;129:1249–1259. - PubMed
-
- Bonora E, Porcelli AM, Gasparre G, Biondi A, Ghelli A, Carelli V, Baracca A, Tallini G, Martinuzzi A, Lenaz G, Rugolo M, Romeo G. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer Res. 2006;66:6087–6096. - PubMed
-
- Brookes PS, Pinner A, Ramachandran A, Coward L, Barnes S, Kim H, Darley-Usmar VM. High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signaling complexes. Proteomics. 2002;2:969–977. - PubMed
-
- Budde SM, van den Heuvel LP, Janssen AJ, Smeets RJ, Buskens CA, DeMeirleir L, Van Coster R, Baethmann M, Voit T, Trijbels JM, Smeitink JA. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem Biophys Res Commun. 2000;275:63–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

