Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis
- PMID: 19461889
- PMCID: PMC2679201
- DOI: 10.1371/journal.pgen.1000492
Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis
Abstract
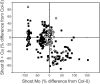
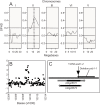
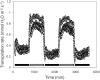
Though central to our understanding of how roots perform their vital function of scavenging water and solutes from the soil, no direct genetic evidence currently exists to support the foundational model that suberin acts to form a chemical barrier limiting the extracellular, or apoplastic, transport of water and solutes in plant roots. Using the newly characterized enhanced suberin1 (esb1) mutant, we established a connection in Arabidopsis thaliana between suberin in the root and both water movement through the plant and solute accumulation in the shoot. Esb1 mutants, characterized by increased root suberin, were found to have reduced day time transpiration rates and increased water-use efficiency during their vegetative growth period. Furthermore, these changes in suberin and water transport were associated with decreases in the accumulation of Ca, Mn, and Zn and increases in the accumulation of Na, S, K, As, Se, and Mo in the shoot. Here, we present direct genetic evidence establishing that suberin in the roots plays a critical role in controlling both water and mineral ion uptake and transport to the leaves. The changes observed in the elemental accumulation in leaves are also interpreted as evidence that a significant component of the radial root transport of Ca, Mn, and Zn occurs in the apoplast.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






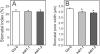


References
-
- Hose E, Clarkson DT, Steudle E, Schreiber L, Hartung W. The exodermis: a variable apoplastic barrier. J Exp Bot. 2001;52:2245–2264. - PubMed
-
- Enstone DE, Peterson CA, Ma FS. Root endodermis and exodermis: Structure, function, and responses to the environment. J Plant Growth Regul. 2002;21:335–351.
-
- Zimmermann HM, Hartmann K, Schreiber L, Steudle E. Chemical composition of apoplastic transport barriers in relation to radial hydraulic conductivity of corn roots (Zea mays L.). Planta. 2000;210:302–311. - PubMed
-
- Freundl E, Steudle E, Hartung W. Apoplastic transport of abscisic acid through roots of maize: effect of the exodermis. Planta. 2000;210:222–231. - PubMed
-
- Harrison-Murray RS, Clarkson DT. Relationships between structural development and absorption of ions by root system of Cucurbita-Pepo. Planta. 1973;114:1–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

