Mapping all-atom models onto one-bead Coarse Grained Models: general properties and applications to a minimal polypeptide model
- PMID: 19461947
- PMCID: PMC2600716
- DOI: 10.1021/ct050294k
Mapping all-atom models onto one-bead Coarse Grained Models: general properties and applications to a minimal polypeptide model
Abstract
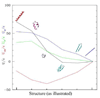
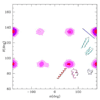
In the one and two beads Coarse Grained (CG) models for proteins, the two conformational dihedrals ϕ and ψ that describe the backbone geometry are no longer present as explicit internal coordinates, thus the information contained in the Ramachandran plot cannot be used directly. We derive an analytical mapping between these dihedrals and the internal variable describing the backbone conformation in the one(two) beads CG models, namely the pseudo-bond angle and pseudo-dihedral between subsequent Cαs. This is used to derive a new density plot that contains the same information as the Ramachandran plot and can be used with the one(two) beads CG models. The use of this mapping is then illustrated with a new one bead polypeptide model that accounts for transitions between α-helices and β-sheets.
Figures






Similar articles
-
Coarse-Grained Potentials for Local Interactions in Unfolded Proteins.J Chem Theory Comput. 2013 Jan 8;9(1):432-40. doi: 10.1021/ct300684j. Epub 2012 Nov 16. J Chem Theory Comput. 2013. PMID: 26589045
-
Revisiting the Ramachandran plot from a new angle.Protein Sci. 2011 Jul;20(7):1166-71. doi: 10.1002/pro.644. Epub 2011 May 31. Protein Sci. 2011. PMID: 21538644 Free PMC article.
-
Separation of time scale and coupling in the motion governed by the coarse-grained and fine degrees of freedom in a polypeptide backbone.J Chem Phys. 2007 Oct 21;127(15):155103. doi: 10.1063/1.2784200. J Chem Phys. 2007. PMID: 17949219
-
Adaptive resolution simulations of biomolecular systems.Eur Biophys J. 2017 Dec;46(8):821-835. doi: 10.1007/s00249-017-1248-0. Epub 2017 Sep 13. Eur Biophys J. 2017. PMID: 28905203 Review.
-
Modeling of Protein Structural Flexibility and Large-Scale Dynamics: Coarse-Grained Simulations and Elastic Network Models.Int J Mol Sci. 2018 Nov 6;19(11):3496. doi: 10.3390/ijms19113496. Int J Mol Sci. 2018. PMID: 30404229 Free PMC article. Review.
Cited by
-
Coiled-coil domains are sufficient to drive liquid-liquid phase separation in protein models.Biophys J. 2024 Mar 19;123(6):703-717. doi: 10.1016/j.bpj.2024.02.007. Epub 2024 Feb 15. Biophys J. 2024. PMID: 38356260 Free PMC article.
-
Multiscale coarse-graining of the protein energy landscape.PLoS Comput Biol. 2010 Jun 24;6(6):e1000827. doi: 10.1371/journal.pcbi.1000827. PLoS Comput Biol. 2010. PMID: 20585614 Free PMC article.
-
Nonspecific interactions can lead to liquid-liquid phase separation in coiled-coil proteins models.bioRxiv [Preprint]. 2025 May 15:2025.05.09.653163. doi: 10.1101/2025.05.09.653163. bioRxiv. 2025. PMID: 40463193 Free PMC article. Preprint.
-
Improving Internal Peptide Dynamics in the Coarse-Grained MARTINI Model: Toward Large-Scale Simulations of Amyloid- and Elastin-like Peptides.J Chem Theory Comput. 2012 May 8;8(5):1774-1785. doi: 10.1021/ct200876v. Epub 2012 Mar 26. J Chem Theory Comput. 2012. PMID: 22582033 Free PMC article.
-
Structural Transition States Explored With Minimalist Coarse Grained Models: Applications to Calmodulin.Front Mol Biosci. 2019 Oct 15;6:104. doi: 10.3389/fmolb.2019.00104. eCollection 2019. Front Mol Biosci. 2019. PMID: 31750313 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
