Characterizing gene family evolution
- PMID: 19461954
- PMCID: PMC2683547
- DOI: 10.1251/bpo144
Characterizing gene family evolution
Abstract
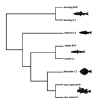
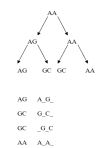
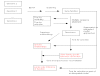
Gene families are widely used in comparative genomics, molecular evolution, and in systematics. However, they are constructed in different manners, their data analyzed and interpreted differently, with different underlying assumptions, leading to sometimes divergent conclusions. In systematics, concepts like monophyly and the dichotomy between homoplasy and homology have been central to the analysis of phylogenies. We critique the traditional use of such concepts as applied to gene families and give examples of incorrect inferences they may lead to. Operational definitions that have emerged within functional genomics are contrasted with the common formal definitions derived from systematics. Lastly, we question the utility of layers of homology and the meaning of homology at the character state level in the context of sequence evolution. From this, we move forward to present an idealized strategy for characterizing gene family evolution for both systematic and functional purposes, including recent methodological improvements.
Keywords: evolution, molecular; genomics; phylogeny; sequence homology.
Figures
Similar articles
-
Homoplasy and homology: dichotomy or continuum?J Hum Evol. 2007 May;52(5):473-9. doi: 10.1016/j.jhevol.2006.11.010. Epub 2007 Feb 20. J Hum Evol. 2007. PMID: 17434571 Review.
-
Gene family evolution and homology: genomics meets phylogenetics.Annu Rev Genomics Hum Genet. 2000;1:41-73. doi: 10.1146/annurev.genom.1.1.41. Annu Rev Genomics Hum Genet. 2000. PMID: 11701624 Review.
-
Advances in Fungal Phylogenomics and Their Impact on Fungal Systematics.Adv Genet. 2017;100:309-328. doi: 10.1016/bs.adgen.2017.09.004. Epub 2017 Oct 20. Adv Genet. 2017. PMID: 29153403
-
Homologues and homology and their related terms in phylogenetic systematics.Cladistics. 2023 Jun;39(3):240-248. doi: 10.1111/cla.12526. Epub 2023 Feb 4. Cladistics. 2023. PMID: 36738298
-
Characterizing lineage-specific evolution and the processes driving genomic diversification in chordates.BMC Evol Biol. 2020 Feb 11;20(1):24. doi: 10.1186/s12862-020-1585-y. BMC Evol Biol. 2020. PMID: 32046633 Free PMC article.
Cited by
-
An Efficient Feature Selection Algorithm for Gene Families Using NMF and ReliefF.Genes (Basel). 2023 Feb 6;14(2):421. doi: 10.3390/genes14020421. Genes (Basel). 2023. PMID: 36833348 Free PMC article.
-
Genome-wide characterization and expression profiling of FARL (FHY3/FAR1) family genes in Zea mays.J Genet Eng Biotechnol. 2024 Sep;22(3):100401. doi: 10.1016/j.jgeb.2024.100401. Epub 2024 Jul 31. J Genet Eng Biotechnol. 2024. PMID: 39179323 Free PMC article.
-
Genomic Survey of Tyrosine Kinases Repertoire in Electrophorus electricus With an Emphasis on Evolutionary Conservation and Diversification.Evol Bioinform Online. 2020 May 25;16:1176934320922519. doi: 10.1177/1176934320922519. eCollection 2020. Evol Bioinform Online. 2020. PMID: 32546936 Free PMC article.
-
The salamander blastema within the broader context of metazoan regeneration.Front Cell Dev Biol. 2023 Aug 11;11:1206157. doi: 10.3389/fcell.2023.1206157. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37635872 Free PMC article. Review.
References
-
- Massey SE, Churbanov A, Rastogi S, Liberles DA. Characterizing positive and negative selection and their phylogenetic effects. Gene. 2008;418:22–26. - PubMed
-
- Russell RB, Sasieni PD, Sternberg MJ. Supersites within superfolds. Binding site similarity in the absence of homology. J Mol Biol. 1998;282:903–918. - PubMed
-
- Fitch WM. Homology: A personal view on some of the problems. Trends in Genetics. 2000;16:227–231. - PubMed
-
- Hennig W. Urbana, IL: University of Illinois Press; 1979. Phylogenetic systematics.
LinkOut - more resources
Full Text Sources

