Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples
- PMID: 19461970
- PMCID: PMC2680989
- DOI: 10.1371/journal.pone.0005645
Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples
Abstract
Background: Several preprocessing algorithms for Affymetrix gene expression microarrays have been developed, and their performance on spike-in data sets has been evaluated previously. However, a comprehensive comparison of preprocessing algorithms on samples taken under research conditions has not been performed.
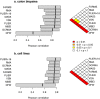
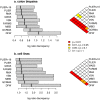
Methodology/principal findings: We used TaqMan RT-PCR arrays as a reference to evaluate the accuracy of expression values from Affymetrix microarrays in two experimental data sets: one comprising 84 genes in 36 colon biopsies, and the other comprising 75 genes in 29 cancer cell lines. We evaluated consistency using the Pearson correlation between measurements obtained on the two platforms. Also, we introduce the log-ratio discrepancy as a more relevant measure of discordance between gene expression platforms. Of nine preprocessing algorithms tested, PLIER+16 produced expression values that were most consistent with RT-PCR measurements, although the difference in performance between most of the algorithms was not statistically significant.
Conclusions/significance: Our results support the choice of PLIER+16 for the preprocessing of clinical Affymetrix microarray data. However, other algorithms performed similarly and are probably also good choices.
Conflict of interest statement
Figures
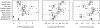
Similar articles
-
Probe set filtering increases correlation between Affymetrix GeneChip and qRT-PCR expression measurements.BMC Bioinformatics. 2010 Feb 24;11:104. doi: 10.1186/1471-2105-11-104. BMC Bioinformatics. 2010. PMID: 20181266 Free PMC article.
-
Comparing the use of Affymetrix to spotted oligonucleotide microarrays using two retinal pigment epithelium cell lines.Mol Vis. 2003 Oct 6;9:482-96. Mol Vis. 2003. PMID: 14551534 Free PMC article.
-
Cross-platform comparison of SYBR Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study.BMC Genomics. 2008 Jul 11;9:328. doi: 10.1186/1471-2164-9-328. BMC Genomics. 2008. PMID: 18620571 Free PMC article.
-
Validation of oligonucleotide microarray data using microfluidic low-density arrays: a new statistical method to normalize real-time RT-PCR data.Biotechniques. 2005 May;38(5):785-92. doi: 10.2144/05385MT01. Biotechniques. 2005. PMID: 15945375
-
Statistical challenges in preprocessing in microarray experiments in cancer.Clin Cancer Res. 2008 Oct 1;14(19):5959-66. doi: 10.1158/1078-0432.CCR-07-4532. Clin Cancer Res. 2008. PMID: 18829474 Free PMC article. Review.
Cited by
-
Layered signaling regulatory networks analysis of gene expression involved in malignant tumorigenesis of non-resolving ulcerative colitis via integration of cross-study microarray profiles.PLoS One. 2013 Jun 25;8(6):e67142. doi: 10.1371/journal.pone.0067142. Print 2013. PLoS One. 2013. PMID: 23825635 Free PMC article.
-
Algorithm-driven artifacts in median polish summarization of microarray data.BMC Bioinformatics. 2010 Nov 11;11:553. doi: 10.1186/1471-2105-11-553. BMC Bioinformatics. 2010. PMID: 21070630 Free PMC article.
-
Long Noncoding RNA PCAT6 Regulates Cell Proliferation and Migration in Human Esophageal Squamous Cell Carcinoma.J Cancer. 2022 Jan 1;13(2):681-690. doi: 10.7150/jca.62671. eCollection 2022. J Cancer. 2022. PMID: 35069911 Free PMC article.
-
Identification of four novel hub genes as monitoring biomarkers for colorectal cancer.Hereditas. 2022 Jan 29;159(1):11. doi: 10.1186/s41065-021-00216-7. Hereditas. 2022. PMID: 35093172 Free PMC article.
-
Pleiotropic functions of EAPII/TTRAP/TDP2: cancer development, chemoresistance and beyond.Cell Cycle. 2011 Oct 1;10(19):3274-83. doi: 10.4161/cc.10.19.17763. Epub 2011 Oct 1. Cell Cycle. 2011. PMID: 21926483 Free PMC article. Review.
References
-
- Affymetrix. 2008 Latin Square Data [ http://www.affymetrix.com/support/technical/sample_data/datasets.affx]
-
- Cope LM, Irizarry RA, Jaffee HA, Wu ZJ, Speed TP. A benchmark for affymetrix GeneChip expression measures. Bioinformatics. 2004;20:323–331. - PubMed
-
- Tong W, Lucas AB, Shippy R, Fan X, Fang H, et al. Evaluation of external RNA controls for the assessment of microarray performance. Nat Biotechnol. 2006;24:1132–1139. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

