Molecular profiling of the "plexinome" in melanoma and pancreatic cancer
- PMID: 19462467
- PMCID: PMC2989154
- DOI: 10.1002/humu.21017
Molecular profiling of the "plexinome" in melanoma and pancreatic cancer
Abstract
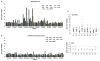
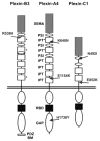
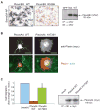
Plexins are transmembrane high-affinity receptors for semaphorins, regulating cell guidance, motility, and invasion. Functional evidences implicate semaphorin signals in cancer progression and metastasis. Yet, it is largely unknown whether plexin genes are genetically altered in human tumors. We performed a comprehensive gene copy analysis and mutational profiling of all nine members of the plexin gene family (plexinome), in melanomas and pancreatic ductal adenocarcinomas (PDACs), which are characterized by high metastatic potential and poor prognosis. Gene copy analysis detected amplification of PLXNA4 in melanomas, whereas copy number losses of multiple plexin genes were seen in PDACs. Somatic mutations were detected in PLXNA4, PLXNB3, and PLXNC1; providing the first evidence that these plexins are mutated in human cancer. Functional assays in cellular models revealed that some of these missense mutations result in loss of plexin function. For instance, c.1613G>A, p.R538H mutation in the extracellular domain of PLXNB3 prevented binding of the ligand Sema5A. Moreover, although PLXNA4 signaling can inhibit tumor cell migration, the mutated c.5206C>T, p.H1736Y allele had lost this activity. Our study is the first systematic analysis of the "plexinome" in human tumors, and indicates that multiple mutated plexins may be involved in cancer progression.
Figures

References
-
- Balakrishnan A, Bleeker FE, Lamba S, Rodolfo M, Daniotti M, Scarpa A, van Tilborg AA, Leenstra S, Zanon C, Bardelli A. Novel somatic and germline mutations in cancer candidate genes in glioblastoma, melanoma, and pancreatic carcinoma. Cancer Res. 2007;67:3545–3550. - PubMed
-
- Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, Cascinelli N, Urist M, McMasters KM, Ross MI, Kirkwood JM, Atkins MB, Thompson JA, Coit DG, Byrd D, Desmond R, Zhang Y, Liu PY, Lyman GH, Morabito A. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19:3622–3634. - PubMed
-
- Barberis D, Artigiani S, Casazza A, Corso S, Giordano S, Love CA, Jones EY, Comoglio PM, Tamagnone L. Plexin signaling hampers integrin-based adhesion, leading to Rho-kinase independent cell rounding, and inhibiting lamellipodia extension and cell motility. Faseb J. 2004;18:592–594. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

