Extensive molecular differences between anterior- and posterior-half-sclerotomes underlie somite polarity and spinal nerve segmentation
- PMID: 19463158
- PMCID: PMC2693541
- DOI: 10.1186/1471-213X-9-30
Extensive molecular differences between anterior- and posterior-half-sclerotomes underlie somite polarity and spinal nerve segmentation
Abstract
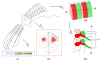
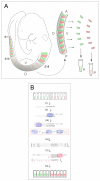
Background: The polarization of somite-derived sclerotomes into anterior and posterior halves underlies vertebral morphogenesis and spinal nerve segmentation. To characterize the full extent of molecular differences that underlie this polarity, we have undertaken a systematic comparison of gene expression between the two sclerotome halves in the mouse embryo.
Results: Several hundred genes are differentially-expressed between the two sclerotome halves, showing that a marked degree of molecular heterogeneity underpins the development of somite polarity.
Conclusion: We have identified a set of genes that warrant further investigation as regulators of somite polarity and vertebral morphogenesis, as well as repellents of spinal axon growth. Moreover the results indicate that, unlike the posterior half-sclerotome, the central region of the anterior-half-sclerotome does not contribute bone and cartilage to the vertebral column, being associated instead with the development of the segmented spinal nerves.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

