Phylogenetic classification of Escherichia coli O157:H7 strains of human and bovine origin using a novel set of nucleotide polymorphisms
- PMID: 19463166
- PMCID: PMC2718522
- DOI: 10.1186/gb-2009-10-5-r56
Phylogenetic classification of Escherichia coli O157:H7 strains of human and bovine origin using a novel set of nucleotide polymorphisms
Abstract
Background: Cattle are a reservoir of Shiga toxin-producing Escherichia coli O157:H7 (STEC O157), and are known to harbor subtypes not typically found in clinically ill humans. Consequently, nucleotide polymorphisms previously discovered via strains originating from human outbreaks may be restricted in their ability to distinguish STEC O157 genetic subtypes present in cattle. The objectives of this study were firstly to identify nucleotide polymorphisms in a diverse sampling of human and bovine STEC O157 strains, secondly to classify strains of either bovine or human origin by polymorphism-derived genotypes, and finally to compare the genotype diversity with pulsed-field gel electrophoresis (PFGE), a method currently used for assessing STEC O157 diversity.
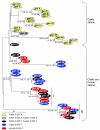
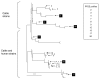
Results: High-throughput 454 sequencing of pooled STEC O157 strain DNAs from human clinical cases (n = 91) and cattle (n = 102) identified 16,218 putative polymorphisms. From those, 178 were selected primarily within genomic regions conserved across E. coli serotypes and genotyped in 261 STEC O157 strains. Forty-two unique genotypes were observed that are tagged by a minimal set of 32 polymorphisms. Phylogenetic trees of the genotypes are divided into clades that represent strains of cattle origin, or cattle and human origin. Although PFGE diversity surpassed genotype diversity overall, ten PFGE patterns each occurred with multiple strains having different genotypes.
Conclusions: Deep sequencing of pooled STEC O157 DNAs proved highly effective in polymorphism discovery. A polymorphism set has been identified that characterizes genetic diversity within STEC O157 strains of bovine origin, and a subset observed in human strains. The set may complement current techniques used to classify strains implicated in disease outbreaks.
Figures



References
-
- Griffin PM, Tauxe RV. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. - PubMed
-
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. - DOI - PubMed
-
- Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

