Gene expression during Drosophila melanogaster egg development before and after reproductive diapause
- PMID: 19463195
- PMCID: PMC2700134
- DOI: 10.1186/1471-2164-10-242
Gene expression during Drosophila melanogaster egg development before and after reproductive diapause
Abstract
Background: Despite the importance of egg development to the female life cycle in Drosophila, global patterns of gene expression have not been examined in detail, primarily due to the difficulty in isolating synchronised developmental stages in sufficient quantities for gene expression profiling. Entry into vitellogenesis is a key stage of oogenesis and by forcing females into reproductive diapause we are able to arrest oogenesis at the pre-vitellogenic stages. Releasing females from diapause allows collection of relatively synchronous developing egg populations and an investigation of some of the transcriptional dynamics apparent before and after reproductive diapause.
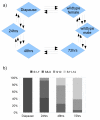
Results: Focusing on gender-biased transcription, we identified mechanisms of egg development suppressed during reproductive dormancy as well as other molecular changes unique to the diapausing female. A microarray based analysis generated a set of 3565 transcripts with at least 2-fold greater expression in females as compared to control males, 1392 such changes were biased during reproductive dormancy. In addition, we also detect 1922 up-regulated transcriptional changes after entry into vitellogenesis, which were classified into discrete blocks of co-expression. We discuss some of the regulatory aspects apparent after re-initiation of egg development, exploring the underlying functions, maternal contribution and evolutionary conservation of co-expression patterns involved in egg production.
Conclusion: Although much of the work we present is descriptive, fundamental aspects of egg development and gender-biased transcription can be derived from our time-series experiment. We believe that our dataset will facilitate further exploration of the developmental and evolutionary characteristics of oogenesis as well as the nature of reproductive arrest in Drosophila.
Figures



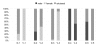
References
-
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. - PubMed
-
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, Bussemaker HJ, White KP. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. - PubMed
-
- Spradling AC. Developmental Genetics of Oogenesis. In: Bate M, Arias AM, editor. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; 1993. pp. 1–70.
-
- Mahowald AP, Kambysellis MP. Oogenesis. In: Ashburner M, Wright TRW, editor. The Genetics and Biology of Drosophila. 2. Academic; 1980. pp. 141–224.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

