Molecular characterization of a new PToV strain. Evolutionary implications
- PMID: 19463719
- PMCID: PMC7114482
- DOI: 10.1016/j.virusres.2009.02.019
Molecular characterization of a new PToV strain. Evolutionary implications
Abstract
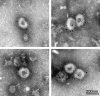
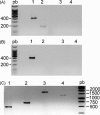
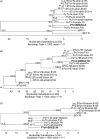
Toroviruses are emergent viruses, belonging to the Nidovirales order, that remain mostly ignored, despite they are able to infect different species of domestic animals and humans, causing enteric diseases and diarrhea. Thus far, only five variants of porcine torovirus (PToV) have been identified. In this report we describe the identification and partial characterization of a new strain of porcine torovirus (PToV-BRES) that was detected by RT-PCR in a swine faecal specimen from a farm in Brescia (Italy). The complete genes coding for the nucleocapsid (N), hemagglutinin-esterase (HE) and membrane (M) proteins were amplified, and sequence analysis showed that PToV-BRES is a new PToV strain that, based on the HE gene sequence, is phylogenetically related to P4 strain, that was up to now the only member of a distinct PToV lineage. The nucleocapsid protein from PToV-BRES was expressed in insect cells as a his-tagged protein, purified by affinity chromatography and used to develop an ELISA method to detect antibodies against PToV. This assay was evaluated using a serum collection including 45 samples from three commercial farms from Spain. High antibody prevalence against PToV was observed in the three farms, both in adult animals and in piglets, which could suggest that PToV might be endemic in Spanish porcine population. The ELISA method developed in this work could be useful in future epidemiological surveys about toroviruses.
Figures


References
-
- Bogdanova V.S., Tsibezov V.V., Grabovetskii V.V., Eliseeva O.V., Grebennikova T.V., Verkhovskii O.A., Zabereshchnyi A.D., Aliper T.I. Enzyme-linked immunosorbent assay for detection of antibodies to porcine reproductive and respiratory syndrome virus, by using recombinant nucleocapsid protein N. Vopr. Virusol. 2007;52(2):45–49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

