Replication and conjugative mobilization of broad host-range IncQ plasmids
- PMID: 19465049
- PMCID: PMC2752045
- DOI: 10.1016/j.plasmid.2009.05.001
Replication and conjugative mobilization of broad host-range IncQ plasmids
Abstract
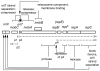
The IncQ plasmids have a broader host-range than any other known replicating element in bacteria. Studies on the replication and conjugative mobilization of these plasmids, which have mostly been focused on the nearly identical RSF1010 and R1162, are summarized with a view to understanding how this broad host-range is achieved. Several significant features of IncQ plasmids emerge from these studies: (1) initiation of replication, involving DnaA-independent activation of the origin and a dedicated primase, is strictly host-independent. (2) The plasmids can be conjugatively mobilized by a variety of different type IV transporters, including those engaged in the secretion of proteins involved in pathogenesis. (3) Stability is insured by a combination of high copy-number and modulated gene expression to reduce metabolic load.
Figures





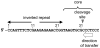
Similar articles
-
DNA replication of IncQ broad-host-range plasmids in gram-negative bacteria.Biosci Biotechnol Biochem. 1996 Mar;60(3):377-82. doi: 10.1271/bbb.60.377. Biosci Biotechnol Biochem. 1996. PMID: 8901094 Review.
-
The primase of broad-host-range plasmid R1162 is active in conjugal transfer.J Bacteriol. 1996 Dec;178(23):6888-94. doi: 10.1128/jb.178.23.6888-6894.1996. J Bacteriol. 1996. PMID: 8955311 Free PMC article.
-
The complete nucleotide sequence and environmental distribution of the cryptic, conjugative, broad-host-range plasmid pIPO2 isolated from bacteria of the wheat rhizosphere.Microbiology (Reading). 2002 Jun;148(Pt 6):1637-1653. doi: 10.1099/00221287-148-6-1637. Microbiology (Reading). 2002. PMID: 12055285
-
Analysis of the mobilization region of the broad-host-range IncQ-like plasmid pTC-F14 and its ability to interact with a related plasmid, pTF-FC2.J Bacteriol. 2003 Oct;185(20):6104-11. doi: 10.1128/JB.185.20.6104-6111.2003. J Bacteriol. 2003. PMID: 14526022 Free PMC article.
-
Replication of plasmids in gram-negative bacteria.Microbiol Rev. 1989 Dec;53(4):491-516. doi: 10.1128/mr.53.4.491-516.1989. Microbiol Rev. 1989. PMID: 2687680 Free PMC article. Review.
Cited by
-
The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes.Nucleic Acids Res. 2013 Jan;41(Database issue):D666-75. doi: 10.1093/nar/gks1119. Epub 2012 Nov 23. Nucleic Acids Res. 2013. PMID: 23180763 Free PMC article.
-
CRISPR-Cas systems preferentially target the leading regions of MOBF conjugative plasmids.RNA Biol. 2013 May;10(5):749-61. doi: 10.4161/rna.24202. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535265 Free PMC article.
-
The Mobilizable Plasmid P3 of Salmonella enterica Serovar Typhimurium SL1344 Depends on the P2 Plasmid for Conjugative Transfer into a Broad Range of Bacteria In Vitro and In Vivo.J Bacteriol. 2022 Dec 20;204(12):e0034722. doi: 10.1128/jb.00347-22. Epub 2022 Nov 16. J Bacteriol. 2022. PMID: 36383016 Free PMC article.
-
High Prevalence of Plasmid-Mediated Quinolone Resistance and IncQ Plasmids Carrying qnrS2 Gene in Bacteria from Rivers near Hospitals and Aquaculture in China.PLoS One. 2016 Jul 18;11(7):e0159418. doi: 10.1371/journal.pone.0159418. eCollection 2016. PLoS One. 2016. PMID: 27427763 Free PMC article.
-
Improving a Synechocystis-based photoautotrophic chassis through systematic genome mapping and validation of neutral sites.DNA Res. 2015 Dec;22(6):425-37. doi: 10.1093/dnares/dsv024. Epub 2015 Oct 21. DNA Res. 2015. PMID: 26490728 Free PMC article.
References
-
- Bagdasarian M, Lurz R, Rückert B, Franklin FC, Bagdasarian MM, Frey J, Timmis KN. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. - PubMed
-
- Bagdasarian M, Bagdasarian M, Lurz R, Nordheim A, Frey J, Timmis K. Molecular and functional analysis of the broad host range plasmid RSF1010 and construction of vectors for gene cloning in gram-negative bacteria. In: Mitsuhashi S, editor. Drug Resistance in Bacteria. New York: Thieme-Stratton; 1982. pp. 183–197.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

