ToppGene Suite for gene list enrichment analysis and candidate gene prioritization
- PMID: 19465376
- PMCID: PMC2703978
- DOI: 10.1093/nar/gkp427
ToppGene Suite for gene list enrichment analysis and candidate gene prioritization
Abstract
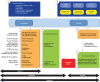
ToppGene Suite (http://toppgene.cchmc.org; this web site is free and open to all users and does not require a login to access) is a one-stop portal for (i) gene list functional enrichment, (ii) candidate gene prioritization using either functional annotations or network analysis and (iii) identification and prioritization of novel disease candidate genes in the interactome. Functional annotation-based disease candidate gene prioritization uses a fuzzy-based similarity measure to compute the similarity between any two genes based on semantic annotations. The similarity scores from individual features are combined into an overall score using statistical meta-analysis. A P-value of each annotation of a test gene is derived by random sampling of the whole genome. The protein-protein interaction network (PPIN)-based disease candidate gene prioritization uses social and Web networks analysis algorithms (extended versions of the PageRank and HITS algorithms, and the K-Step Markov method). We demonstrate the utility of ToppGene Suite using 20 recently reported GWAS-based gene-disease associations (including novel disease genes) representing five diseases. ToppGene ranked 19 of 20 (95%) candidate genes within the top 20%, while ToppNet ranked 12 of 16 (75%) candidate genes among the top 20%.
Figures
References
-
- Freudenberg J, Propping P. A similarity-based method for genome-wide prediction of disease-relevant human genes. Bioinformatics. 2002;18(Suppl. 2):S110–S115. - PubMed
-
- Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent LC, De Moor B, Marynen P, Hassan B, et al. Gene prioritization through genomic data fusion. Nat. Biotechnol. 2006;24:537–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

