HotSpot Wizard: a web server for identification of hot spots in protein engineering
- PMID: 19465397
- PMCID: PMC2703904
- DOI: 10.1093/nar/gkp410
HotSpot Wizard: a web server for identification of hot spots in protein engineering
Abstract
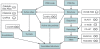
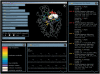
HotSpot Wizard is a web server for automatic identification of 'hot spots' for engineering of substrate specificity, activity or enantioselectivity of enzymes and for annotation of protein structures. The web server implements the protein engineering protocol, which targets evolutionarily variable amino acid positions located in the active site or lining the access tunnels. The 'hot spots' for mutagenesis are selected through the integration of structural, functional and evolutionary information obtained from: (i) the databases RCSB PDB, UniProt, PDBSWS, Catalytic Site Atlas and nr NCBI and (ii) the tools CASTp, CAVER, BLAST, CD-HIT, MUSCLE and Rate4Site. The protein structure and e-mail address are the only obligatory inputs for the calculation. In the output, HotSpot Wizard lists annotated residues ordered by estimated mutability. The results of the analysis are mapped on the enzyme structure and visualized in the web browser using Jmol. The HotSpot Wizard server should be useful for protein engineers interested in exploring the structure of their favourite protein and for the design of mutations in site-directed mutagenesis and focused directed evolution experiments. HotSpot Wizard is available at http://loschmidt.chemi.muni.cz/hotspotwizard/.
Figures

References
-
- Bornscheuer UT, Pohl M. Improved biocatalysts by directed evolution and rational protein design. Curr. Opin. Chem. Biol. 2001;5:137–143. - PubMed
-
- Brannigan JA, Wilkinson AJ. Protein engineering 20 years on. Nat. Rev. Mol. Cell Biol. 2002;3:964–970. - PubMed
-
- Chen R. Enzyme engineering: rational redesign versus directed evolution. Trends Biotechnol. 2001;19:13–14. - PubMed
-
- Chica RA, Doucet N, Pelletier JN. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol. 2005;16:378–384. - PubMed
-
- Park S, Morley KL, Horsman GP, Holmquist M, Hult K, Kazlauskas RJ. Focusing mutations into the P. fluorescens esterase binding site increases enantioselectivity more effectively than distant mutations. Chem. Biol. 2005;12:45–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

