Genome-wide and fine-resolution association analysis of malaria in West Africa
- PMID: 19465909
- PMCID: PMC2889040
- DOI: 10.1038/ng.388
Genome-wide and fine-resolution association analysis of malaria in West Africa
Abstract
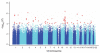
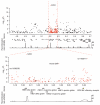
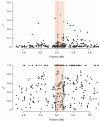
We report a genome-wide association (GWA) study of severe malaria in The Gambia. The initial GWA scan included 2,500 children genotyped on the Affymetrix 500K GeneChip, and a replication study included 3,400 children. We used this to examine the performance of GWA methods in Africa. We found considerable population stratification, and also that signals of association at known malaria resistance loci were greatly attenuated owing to weak linkage disequilibrium (LD). To investigate possible solutions to the problem of low LD, we focused on the HbS locus, sequencing this region of the genome in 62 Gambian individuals and then using these data to conduct multipoint imputation in the GWA samples. This increased the signal of association, from P = 4 × 10(-7) to P = 4 × 10(-14), with the peak of the signal located precisely at the HbS causal variant. Our findings provide proof of principle that fine-resolution multipoint imputation, based on population-specific sequencing data, can substantially boost authentic GWA signals and enable fine mapping of causal variants in African populations.
Figures

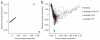
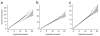
References
-
- Greenwood BM, et al. Mortality and morbidity from malaria among children in a rural area of The Gambia, West Africa. Trans. R. Soc. Trop. Med. Hyg. 1987;81:478–486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U190081993/MRC_/Medical Research Council/United Kingdom
- 077011/WT_/Wellcome Trust/United Kingdom
- 077383/Z/05/Z/WT_/Wellcome Trust/United Kingdom
- 076113/WT_/Wellcome Trust/United Kingdom
- G0600230(77610)/MRC_/Medical Research Council/United Kingdom
- G0800759/MRC_/Medical Research Council/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- 064890/WT_/Wellcome Trust/United Kingdom
- U19 AI065683/AI/NIAID NIH HHS/United States
- G9828345/MRC_/Medical Research Council/United Kingdom
- 076934/WT_/Wellcome Trust/United Kingdom
- 072064/WT_/Wellcome Trust/United Kingdom
- 081682/WT_/Wellcome Trust/United Kingdom
- G0600329/MRC_/Medical Research Council/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- G0600230/MRC_/Medical Research Council/United Kingdom
- G19/9/MRC_/Medical Research Council/United Kingdom
- CZB/4/540/CSO_/Chief Scientist Office/United Kingdom
- 089062/WT_/Wellcome Trust/United Kingdom
- 077383/WT_/Wellcome Trust/United Kingdom
- 061858/WT_/Wellcome Trust/United Kingdom
- G0800675/MRC_/Medical Research Council/United Kingdom
- MC_U190081977/MRC_/Medical Research Council/United Kingdom
- G0600718/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

