PosMed (Positional Medline): prioritizing genes with an artificial neural network comprising medical documents to accelerate positional cloning
- PMID: 19468046
- PMCID: PMC2703941
- DOI: 10.1093/nar/gkp384
PosMed (Positional Medline): prioritizing genes with an artificial neural network comprising medical documents to accelerate positional cloning
Abstract
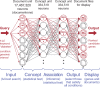
PosMed (http://omicspace.riken.jp/) prioritizes candidate genes for positional cloning by employing our original database search engine GRASE, which uses an inferential process similar to an artificial neural network comprising documental neurons (or 'documentrons') that represent each document contained in databases such as MEDLINE and OMIM. Given a user-specified query, PosMed initially performs a full-text search of each documentron in the first-layer artificial neurons and then calculates the statistical significance of the connections between the hit documentrons and the second-layer artificial neurons representing each gene. When a chromosomal interval(s) is specified, PosMed explores the second-layer and third-layer artificial neurons representing genes within the chromosomal interval by evaluating the combined significance of the connections from the hit documentrons to the genes. PosMed is, therefore, a powerful tool that immediately ranks the candidate genes by connecting phenotypic keywords to the genes through connections representing not only gene-gene interactions but also other biological interactions (e.g. metabolite-gene, mutant mouse-gene, drug-gene, disease-gene and protein-protein interactions) and ortholog data. By utilizing orthologous connections, PosMed facilitates the ranking of human genes based on evidence found in other model species such as mouse. Currently, PosMed, an artificial superbrain that has learned a vast amount of biological knowledge ranging from genomes to phenomes (or 'omic space'), supports the prioritization of positional candidate genes in humans, mouse, rat and Arabidopsis thaliana.
Figures
Similar articles
-
PosMed-plus: an intelligent search engine that inferentially integrates cross-species information resources for molecular breeding of plants.Plant Cell Physiol. 2009 Jul;50(7):1249-59. doi: 10.1093/pcp/pcp086. Epub 2009 Jun 15. Plant Cell Physiol. 2009. PMID: 19528193 Free PMC article.
-
PosMed: Ranking genes and bioresources based on Semantic Web Association Study.Nucleic Acids Res. 2013 Jul;41(Web Server issue):W109-14. doi: 10.1093/nar/gkt474. Epub 2013 Jun 12. Nucleic Acids Res. 2013. PMID: 23761449 Free PMC article.
-
Statistical search on the Semantic Web.Bioinformatics. 2008 Apr 1;24(7):1002-10. doi: 10.1093/bioinformatics/btn054. Epub 2008 Feb 8. Bioinformatics. 2008. PMID: 18263641
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
-
The Riken mouse genome encyclopedia project.C R Biol. 2003 Oct-Nov;326(10-11):923-9. doi: 10.1016/j.crvi.2003.09.018. C R Biol. 2003. PMID: 14744098 Review.
Cited by
-
Candidate gene prioritization.Mol Genet Genomics. 2012 Sep;287(9):679-98. doi: 10.1007/s00438-012-0710-z. Epub 2012 Aug 15. Mol Genet Genomics. 2012. Retraction in: Mol Genet Genomics. 2015 Dec;290(6):2357. doi: 10.1007/s00438-015-1117-4. PMID: 22893106 Retracted. Review.
-
OmicBrowse: a Flash-based high-performance graphics interface for genomic resources.Nucleic Acids Res. 2009 Jul;37(Web Server issue):W57-62. doi: 10.1093/nar/gkp404. Epub 2009 Jun 15. Nucleic Acids Res. 2009. PMID: 19528066 Free PMC article.
-
Gain-of-function mutations in a member of the Src family kinases cause autoinflammatory bone disease in mice and humans.Proc Natl Acad Sci U S A. 2019 Jun 11;116(24):11872-11877. doi: 10.1073/pnas.1819825116. Epub 2019 May 28. Proc Natl Acad Sci U S A. 2019. PMID: 31138708 Free PMC article.
-
Eeyore: a novel mouse model of hereditary deafness.PLoS One. 2013 Sep 23;8(9):e74243. doi: 10.1371/journal.pone.0074243. eCollection 2013. PLoS One. 2013. PMID: 24086324 Free PMC article.
-
A novel gene essential for the development of single positive thymocytes.Mol Cell Biol. 2009 Sep;29(18):5128-35. doi: 10.1128/MCB.00793-09. Epub 2009 Jul 20. Mol Cell Biol. 2009. PMID: 19620281 Free PMC article.
References
-
- Toyoda T, Wada A. Omic space: coordinate-based integration and analysis of genomic phenomic interactions. Bioinformatics. 2004;20:1759–1765. - PubMed
-
- Kobayashi N, Toyoda T. Statistical search on the Semantic Web. Bioinformatics. 2008;24:1002–1010. - PubMed
-
- Prud'hommeaux E, Seaborne A. SPARQL Query Language for RDF, The World Wide Web Consortium, W3C Recommendation 15 January 2008. 2008. http://www.w3.org/TR/2008/REC-rdf-sparql-query-20080115/
-
- Berners-Lee T, Hendler J, Lassila O. The semantic web. Sci. Am. 2001;28:34–43.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

