Non-enzymatic template-directed recombination of RNAs
- PMID: 19468339
- PMCID: PMC2680647
- DOI: 10.3390/ijms10041788
Non-enzymatic template-directed recombination of RNAs
Abstract
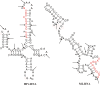
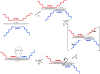
RNA non-enzymatic recombination reactions are of great interest within the hypothesis of the "RNA world", which argues that at some stage of prebiotic life development proteins were not yet engaged in biochemical reactions and RNA carried out both the information storage task and the full range of catalytic roles necessary in primitive self-replicating systems. Here we report on the study of recombination reaction occuring between two 96 nucleotides (nts) fragments of RNAs under physiological conditions and governed by a short oligodeoxyribonucleotide template, partially complementary to sequences within each of the RNAs. Analysis of recombination products shows that ligation is predominantly template-directed, and occurs within the complementary complex with the template in "butt-to-butt" manner, in 1- or 3- nts bulges or in 2-3 nts internal loops. Minor recombination products formed in the template-independent manner are detected as well.
Keywords: RNA; RNA bulge loops; RNA internal loops; RNA world; non-enzymatic ligation; non-enzymatic recombination; origin of life.
Figures




Similar articles
-
Non-enzymatic recombination of RNA: Ligation in loops.Biochim Biophys Acta Gen Subj. 2018 Mar;1862(3):705-725. doi: 10.1016/j.bbagen.2017.10.019. Epub 2017 Oct 31. Biochim Biophys Acta Gen Subj. 2018. PMID: 29097301
-
RNA-RNA recombination in Sindbis virus: roles of the 3' conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching.J Virol. 1997 Apr;71(4):2693-704. doi: 10.1128/JVI.71.4.2693-2704.1997. J Virol. 1997. PMID: 9060622 Free PMC article.
-
Complementarity-directed RNA dimer-linkage promotes retroviral recombination in vivo.Nucleic Acids Res. 2004 Jan 9;32(1):102-14. doi: 10.1093/nar/gkh159. Print 2004. Nucleic Acids Res. 2004. PMID: 14715920 Free PMC article.
-
The puzzle of RNA recombination.FEBS Lett. 1999 Oct 22;460(1):1-5. doi: 10.1016/s0014-5793(99)01282-x. FEBS Lett. 1999. PMID: 10571050 Free PMC article. Review.
-
Genetic reassortment and patch repair by recombination in retroviruses.J Biomed Sci. 2000 Mar-Apr;7(2):77-99. doi: 10.1007/BF02256615. J Biomed Sci. 2000. PMID: 10754383 Review.
Cited by
-
Sex in a test tube: testing the benefits of in vitro recombination.Philos Trans R Soc Lond B Biol Sci. 2016 Oct 19;371(1706):20150529. doi: 10.1098/rstb.2015.0529. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27619693 Free PMC article. Review.
-
Spontaneous advent of genetic diversity in RNA populations through multiple recombination mechanisms.RNA. 2019 Apr;25(4):453-464. doi: 10.1261/rna.068908.118. Epub 2019 Jan 22. RNA. 2019. PMID: 30670484 Free PMC article.
-
Random-sequence genetic oligomer pools display an innate potential for ligation and recombination.Elife. 2018 Nov 21;7:e43022. doi: 10.7554/eLife.43022. Elife. 2018. PMID: 30461419 Free PMC article.
-
Recombination in Enteroviruses, a Multi-Step Modular Evolutionary Process.Viruses. 2019 Sep 14;11(9):859. doi: 10.3390/v11090859. Viruses. 2019. PMID: 31540135 Free PMC article. Review.
References
-
- Gilbert W. The RNA World. Nature. 1986;319:618.
-
- Ertem G, Ferris JP. Formation of RNA oligomers on montmorillonite: site of catalysis. Origins Life Evol. Biosphere. 1998;28:485–499. - PubMed
-
- Ertem G, Ferris JP. Sequence- and regio-selectivity in the montmorillonite-catalyzed synthesis of RNA. Origins Life Evol. Biosphere. 2000;30:411–422. - PubMed
-
- Orgel LE. Some consequences of the RNA world hypothesis. Origins Life Evol. Biosphere. 2003;33:211–218. - PubMed
-
- Sreedhara A, Cowan JA. Structural and catalytic roles for divalent magnesium in nucleic acid biochemistry. Biometals. 2002;15:211–223. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

