Direct observation of the hole protonation state and hole localization site in DNA-oligomers
- PMID: 19469533
- PMCID: PMC2735011
- DOI: 10.1021/ja9014869
Direct observation of the hole protonation state and hole localization site in DNA-oligomers
Abstract
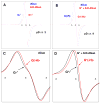
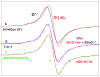
In this work, it is shown that the incorporation of an 8-deuteroguanine (G*) moiety in DNA-oligomers allows for direct determination at 77 K of (i) the location of holes (i.e., the radical site) within dsDNA at specific base sites, even within stacks of G, as well as (ii) the protonation state of the hole at that site. These findings are based on our work and demonstrate that selective deuteration at C-8 on a guanine moiety in dGuo results in an ESR signal from the guanine cation radical (G**(+)) which is easily distinguishable from that of the undeuterated guanine cation radical (G*(+)). G**(+) is also found to be easily distinguishable from its conjugate base, the N1-deprotonated radical, G*(-H)*. Our ESR results clearly establish that at 77 K (i) one-electron oxidized guanine in double stranded DNA-oligomers exists as the deprotonated neutral radical G(-H)* as a result of facile proton transfer to the hydrogen bonded cytosine and (ii) the hole is preferentially located at the 5'-end in several ds DNA-oligomers with a GGG sequence.
Figures


References
-
-
Bernhard WA. Adv Radiat Biol. 1981;9:199–280.Becker D, Sevilla MD. Adv Radiat Biol. 1993;17:121–180.Ward JF. Cold Spring Harb Symp Quant Biol. 2000;65:377–382. and references therein. von Sonntag C. Free-radical-induced DNA Damage and Its Repair. Springer-Verlag; Berlin, Heidelberg: 2006. pp. 335–447.Becker D, Adhikary A, Sevilla MD. In: Charge Migration in DNA: Physics, Chemistry and Biology Perspectives. Chakraborty T, editor. Springer-Verlag; Berlin, Heidelberg, New York: 2007. pp. 139–175.
-
-
- O'Neill MA, Barton JK. In: Charge Transfer in DNA: From Mechanism to Application. Wagenknecht HA, editor. Wiley-VCH Verlag GmbH & Co KGaA; Weiheim: 2005. pp. 27–75.
-
- Schuster GB, editor. Topics in Current Chemistry. Springer-Verlag; Berlin, Heidelberg: 2004. Long Range Charge Transfer in DNA. I and II.
-
- Giese B. Ann Rev Biochem. 2002;71:51–70. - PubMed
-
- Sevilla MD, Becker D, Yan M, Summerfield SR. J Phys Chem. 1991;95:3409–3415.
- Spalletta RA, Bernhard WA. Radiation Research. 1992;130:7–14. - PubMed
- Weiland B, Hüttermann J. Int J Radiat Biol. 1998;74:341–358. - PubMed
- Weiland B, Hüttermann J. Int J Radiat Biol. 1999;75:1169–1175. - PubMed
- Debije MG, Bernhard WA. J Phys Chem B. 2000;104:7845–7851.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

