The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress
- PMID: 19470158
- PMCID: PMC2718523
- DOI: 10.1186/gb-2009-10-5-r57
The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress
Abstract
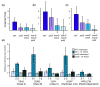
Background: Yeast responding to stress activate a large gene expression program called the Environmental Stress Response that consists of approximately 600 repressed genes and approximately 300 induced genes. Numerous factors are implicated in regulating subsets of Environmental Stress Response genes; however, a complete picture of Environmental Stress Response regulation remains unclear. We investigated the role of the histone deacetylase Rpd3p, previously linked to the upstream regions of many Environmental Stress Response genes, in producing Environmental Stress Response gene expression changes in response to stress.
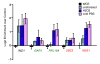
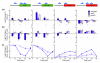
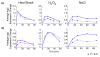
Results: We found that the Rpd3-Large complex is required for proper expression of both induced and repressed Environmental Stress Response genes under multiple stress conditions. Cells lacking RPD3 or the Rpd3-Large subunit PHO23 had a major defect in Environmental Stress Response initiation, particularly during the transient phase of expression immediately after stress exposure. Chromatin-immunoprecipitation showed a direct role for Rpd3-Large at representative genes; however, there were different effects on nucleosome occupancy and histone deacetylation at different promoters. Computational analysis implicated regulators that may act with Rpd3p at Environmental Stress Response genes. We provide genetic and biochemical evidence that Rpd3p is required for binding and action of the stress-activated transcription factor Msn2p, although the contribution of these factors differs for different genes.
Conclusions: Our results implicate Rpd3p as an important co-factor in the Environmental Stress Response regulatory network, and suggest the importance of histone modification in producing transient changes in gene expression triggered by stress.
Figures
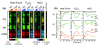
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

