Evaluation of methods for amplification of picogram amounts of total RNA for whole genome expression profiling
- PMID: 19470167
- PMCID: PMC2700135
- DOI: 10.1186/1471-2164-10-246
Evaluation of methods for amplification of picogram amounts of total RNA for whole genome expression profiling
Abstract
Background: For more than a decade, microarrays have been a powerful and widely used tool to explore the transcriptome of biological systems. However, the amount of biological material from cell sorting or laser capture microdissection is much too small to perform microarray studies. To address this issue, RNA amplification methods have been developed to generate sufficient targets from picogram amounts of total RNA to perform microarray hybridisation.
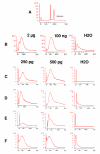
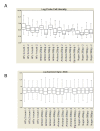
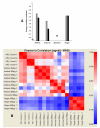
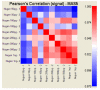
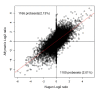
Results: In this study, four commercial protocols for amplification of picograms amounts of input RNA for microarray expression profiling were evaluated and compared. The quantitative and qualitative performances of the methods were assessed. Microarrays were hybridised with the amplified targets and the amplification protocols were compared with respect to the quality of expression profiles, reproducibility within a concentration range of input RNA, and sensitivity. The results demonstrate significant differences between these four methods.
Conclusion: In our hands, the WT-Ovation pico system proposed by Nugen appears to be the most suitable for RNA amplification. This comparative study will be useful to scientists needing to choose an amplification method to carry out microarray experiments involving samples comprising only a few cells and generating picogram amounts of RNA.
Figures
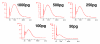
Similar articles
-
Complementary RNA amplification methods enhance microarray identification of transcripts expressed in the C. elegans nervous system.BMC Genomics. 2008 Feb 19;9:84. doi: 10.1186/1471-2164-9-84. BMC Genomics. 2008. PMID: 18284693 Free PMC article.
-
Single-cell laser-capture microdissection and RNA amplification.Methods Mol Med. 2004;99:215-23. doi: 10.1385/1-59259-770-X:215. Methods Mol Med. 2004. PMID: 15131340
-
A comparison of RNA amplification techniques at sub-nanogram input concentration.BMC Genomics. 2009 Jul 20;10:326. doi: 10.1186/1471-2164-10-326. BMC Genomics. 2009. PMID: 19619282 Free PMC article.
-
Strategies for microarray analysis of limiting amounts of RNA.Brief Funct Genomic Proteomic. 2003 Apr;2(1):31-6. doi: 10.1093/bfgp/2.1.31. Brief Funct Genomic Proteomic. 2003. PMID: 15239941 Review.
-
Expression profiling using human tissues in combination with RNA amplification and microarray analysis: assessment of Langerhans cell histiocytosis.Amino Acids. 2005 May;28(3):279-90. doi: 10.1007/s00726-005-0177-x. Epub 2005 Mar 30. Amino Acids. 2005. PMID: 15791395 Review.
Cited by
-
Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model.J Immunol. 2016 Aug 15;197(4):1477-88. doi: 10.4049/jimmunol.1600589. Epub 2016 Jul 1. J Immunol. 2016. PMID: 27371726 Free PMC article.
-
Gene expression differences predict treatment outcome of merkel cell carcinoma patients.J Skin Cancer. 2014;2014:596459. doi: 10.1155/2014/596459. Epub 2014 Jan 30. J Skin Cancer. 2014. PMID: 24634783 Free PMC article.
-
Chemical transfection of dye-conjugated microRNA precursors for microRNA functional analysis of M2 macrophages.J Cell Biochem. 2012 May;113(5):1714-23. doi: 10.1002/jcb.24041. J Cell Biochem. 2012. PMID: 22213010 Free PMC article.
-
Implications of zonal architecture on differential gene expression profiling and altered pathway expressions in mandibular condylar cartilage.Sci Rep. 2021 Aug 19;11(1):16915. doi: 10.1038/s41598-021-96071-7. Sci Rep. 2021. PMID: 34413358 Free PMC article.
-
Whole genome and transcriptome amplification: practicable tools for sustainable tissue biobanking?Virchows Arch. 2012 Nov;461(5):571-80. doi: 10.1007/s00428-012-1315-y. Epub 2012 Sep 25. Virchows Arch. 2012. PMID: 23007645
References
-
- Ibrahim SF, Engh G van den. Flow cytometry and cell sorting. Adv Biochem Eng Biotechnol. 2007;106:19–39. - PubMed
-
- Suarez-Quian CA, Goldstein SR, Pohida T, Smith PD, Peterson JI, Wellner E, Ghany M, Bonner RF. Laser capture microdissection of single cells from complex tissues. Biotechniques. 1999;26:328–335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

