Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites
- PMID: 19470487
- PMCID: PMC2695064
- DOI: 10.1073/pnas.0900153106
Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites
Abstract
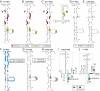
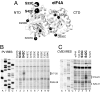
Viral internal ribosomal entry sites (IRESs) mediate end-independent translation initiation. There are 4 major structurally-distinct IRES groups: type 1 (e.g., poliovirus) and type 2 (e.g., encephalomyocarditis virus), which are dissimilar except for a Yn-Xm-AUG motif at their 3' borders, type 3 (e.g., hepatitis C virus), and type 4 (dicistroviruses). Type 2-4 IRESs mediate initiation by distinct mechanisms that are nevertheless all based on specific noncanonical interactions with canonical components of the translation apparatus, such as eukaryotic initiation factor (eIF) 4G (type 2), 40S ribosomal subunits (types 3 and 4), and eIF3 (type 3). The mechanism of initiation on type 1 IRESs is unknown. We now report that domain V of type 1 IRESs, which is adjacent to the Yn-Xm-AUG motif, specifically interacts with the central domain of eIF4G. The position and orientation of eIF4G relative to the Yn-Xm-AUG motif is analogous in type 1 and 2 IRESs. eIF4G promotes recruitment of eIF4A to type 1 IRESs, and together, eIF4G and eIF4A induce conformational changes at their 3' borders. The ability of mutant type 1 IRESs to bind eIF4G/eIF4A correlated with their translational activity. These characteristics parallel the mechanism of initiation on type 2 IRESs, in which the key event is binding of eIF4G to the J-K domain adjacent to the Yn-Xm-AUG motif, which is enhanced by eIF4A. These data suggest that fundamental aspects of the mechanisms of initiation on these unrelated classes of IRESs are similar.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


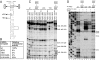
References
-
- Pestova TV, Lorsch J, Hellen CUT. In: Translational Control in Biology and Medicine. Mathews M, Sonenberg N, Hershey J, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 87–120.
-
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. - PubMed
-
- Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol. 2000;20:6019–6029. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

