AdnAB: a new DSB-resecting motor-nuclease from mycobacteria
- PMID: 19470566
- PMCID: PMC2701575
- DOI: 10.1101/gad.1805709
AdnAB: a new DSB-resecting motor-nuclease from mycobacteria
Abstract
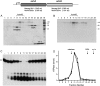
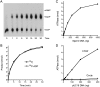
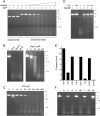
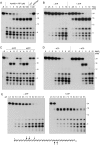
The resection of DNA double-strand breaks (DSBs) in bacteria is a motor-driven process performed by a multisubunit helicase-nuclease complex: either an Escherichia coli-type RecBCD enzyme or a Bacillus-type AddAB enzyme. Here we identify mycobacterial AdnAB as the founder of a new family of heterodimeric helicase-nucleases with distinctive properties. The AdnA and AdnB subunits are each composed of an N-terminal UvrD-like motor domain and a C-terminal nuclease module. The AdnAB ATPase is triggered by dsDNA with free ends and the energy of ATP hydrolysis is coupled to DSB end resection by the AdnAB nuclease. The mycobacterial nonhomologous end-joining (NHEJ) protein Ku protects DSBs from resection by AdnAB. We find that AdnAB incises ssDNA by measuring the distance from the free 5' end to dictate the sites of cleavage, which are predominantly 5 or 6 nucleotides (nt) from the 5' end. The "molecular ruler" of AdnAB is regulated by ATP, which elicits an increase in ssDNA cleavage rate and a distal displacement of the cleavage sites 16-17 nt from the 5' terminus. AdnAB is a dual nuclease with a clear division of labor between the subunits. Mutations in the nuclease active site of the AdnB subunit ablate the ATP-inducible cleavages; the corresponding changes in AdnA abolish ATP-independent cleavage. Complete suppression of DSB end resection requires simultaneous mutation of both subunit nucleases. The nuclease-null AdnAB is a helicase that unwinds linear plasmid DNA without degrading the displaced single strands. Mutations of the phosphohydrolase active site of the AdnB subunit ablate DNA-dependent ATPase activity, DSB end resection, and ATP-inducible ssDNA cleavage; the equivalent mutations of the AdnA subunit have comparatively little effect. AdnAB is a novel signature of the Actinomycetales taxon. Mycobacteria are exceptional in that they encode both AdnAB and RecBCD, suggesting the existence of alternative end-resecting motor-nuclease complexes.
Figures




Comment in
-
Multiplicity of DNA end resection machineries in chromosome break repair.Genes Dev. 2009 Jul 1;23(13):1481-6. doi: 10.1101/gad.1824209. Genes Dev. 2009. PMID: 19571177 Free PMC article.
Similar articles
-
Characterization of the mycobacterial AdnAB DNA motor provides insights into the evolution of bacterial motor-nuclease machines.J Biol Chem. 2010 Jan 22;285(4):2632-41. doi: 10.1074/jbc.M109.076133. Epub 2009 Nov 17. J Biol Chem. 2010. PMID: 19920138 Free PMC article.
-
Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.Proc Natl Acad Sci U S A. 2019 Dec 3;116(49):24507-24516. doi: 10.1073/pnas.1913546116. Epub 2019 Nov 18. Proc Natl Acad Sci U S A. 2019. PMID: 31740608 Free PMC article.
-
Double strand break unwinding and resection by the mycobacterial helicase-nuclease AdnAB in the presence of single strand DNA-binding protein (SSB).J Biol Chem. 2010 Nov 5;285(45):34319-29. doi: 10.1074/jbc.M110.162925. Epub 2010 Aug 23. J Biol Chem. 2010. PMID: 20736178 Free PMC article.
-
Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB.Nat Rev Microbiol. 2013 Jan;11(1):9-13. doi: 10.1038/nrmicro2917. Epub 2012 Dec 3. Nat Rev Microbiol. 2013. PMID: 23202527 Review.
-
Review of DNA repair enzymes in bacteria: With a major focus on AddAB and RecBCD.DNA Repair (Amst). 2022 Oct;118:103389. doi: 10.1016/j.dnarep.2022.103389. Epub 2022 Aug 24. DNA Repair (Amst). 2022. PMID: 36030574 Review.
Cited by
-
DNA end resection--unraveling the tail.DNA Repair (Amst). 2011 Mar 7;10(3):344-8. doi: 10.1016/j.dnarep.2010.12.004. Epub 2011 Jan 11. DNA Repair (Amst). 2011. PMID: 21227759 Free PMC article. Review.
-
Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation.Nucleic Acids Res. 2014;42(17):11056-70. doi: 10.1093/nar/gku752. Epub 2014 Sep 8. Nucleic Acids Res. 2014. PMID: 25200080 Free PMC article.
-
Distributive Conjugal Transfer: New Insights into Horizontal Gene Transfer and Genetic Exchange in Mycobacteria.Microbiol Spectr. 2014;2(1):04. doi: 10.1128/microbiolspec.MGM2-0022-2013. Microbiol Spectr. 2014. PMID: 25505644 Free PMC article.
-
End-resection at DNA double-strand breaks in the three domains of life.Biochem Soc Trans. 2013 Feb 1;41(1):314-20. doi: 10.1042/BST20120307. Biochem Soc Trans. 2013. PMID: 23356304 Free PMC article. Review.
-
Gap filling activities of Pseudomonas DNA ligase D (LigD) polymerase and functional interactions of LigD with the DNA end-binding Ku protein.J Biol Chem. 2010 Feb 12;285(7):4815-25. doi: 10.1074/jbc.M109.073874. Epub 2009 Dec 15. J Biol Chem. 2010. PMID: 20018881 Free PMC article.
References
-
- Chedin F, Kowalczykowski SC. A novel family of regulated helicases/nucleases for Gram-positive bacteria: Insights into the initiation of DNA recombination. Mol Microbiol. 2002;43:823–834. - PubMed
-
- Chen HW, Radle DE, Gabbidon M, Julin DA. Functions of the ATP hydrolysis subunits (RecB and RecD) in the nuclease reactions catalyzed by the RecBCD enzyme from Escherichia coli. J Mol Biol. 1998;278:89–104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
