Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly
- PMID: 19470664
- PMCID: PMC2704428
- DOI: 10.1101/gr.089417.108
Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly
Abstract
Broad applications of zinc finger nuclease (ZFN) technology-which allows targeted genome editing-in research, medicine, and biotechnology are hampered by the lack of a convenient, rapid, and publicly available method for the synthesis of functional ZFNs. Here we describe an efficient and easy-to-practice modular-assembly method using publicly available zinc fingers to make ZFNs that can modify the DNA sequences of predetermined genomic sites in human cells. We synthesized and tested hundreds of ZFNs to target dozens of different sites in the human CCR5 gene-a co-receptor required for HIV infection-and found that many of these nucleases induced site-specific mutations in the CCR5 sequence. Because human cells that harbor CCR5 null mutations are functional and normal, these ZFNs might be used for (1) knocking out CCR5 to produce T-cells that are resistant to HIV infection in AIDS patients or (2) inserting therapeutic genes at "safe sites" in gene therapy applications.
Figures
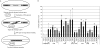

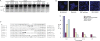
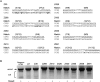
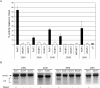
References
-
- Bae KH, Kim JS. One-step selection of artificial transcription factors using an in vivo screening system. Mol Cells. 2006;21:376–380. - PubMed
-
- Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 2003;21:275–280. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
