The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells
- PMID: 19470746
- PMCID: PMC2715673
- DOI: 10.1128/IAI.00096-09
The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells
Abstract
The ability of Acinetobacter baumannii to adhere to and persist on surfaces as biofilms could be central to its pathogenicity. The production of pili and a biofilm-associated protein and the expression of antibiotic resistance are needed for robust biofilm formation on abiotic and biotic surfaces. This multistep process also depends on the expression of transcriptional regulatory functions, some of which could sense nutrients available to cells. This report extends previous observations by showing that although outer membrane protein A (OmpA) of A. baumannii 19606 plays a partial role in the development of robust biofilms on plastic, it is essential for bacterial attachment to Candida albicans filaments and A549 human alveolar epithelial cells. In contrast to abiotic surfaces, the interaction with biotic surfaces is independent of the CsuA/BABCDE-mediated pili. The interaction of A. baumannii 19606 with fungal and epithelial cells also results in their apoptotic death, a response that depends on the direct contact of bacteria with these two types of eukaryotic cells. Furthermore, the bacterial adhesion phenotype correlates with the ability of bacteria to invade A549 epithelial cells. Interestingly, the killing activity of cell-free culture supernatants proved to be protease and temperature sensitive, suggesting that its cytotoxic activity is due to secreted proteins, some of which are different from OmpA.
Figures



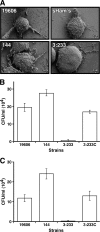
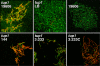
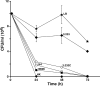
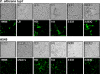

References
-
- Barcak, J. G., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204321-432. - PubMed
-
- Barrios, A. F., R. Zuo, D. Ren, and T. K. Wood. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93188-200. - PubMed
-
- Bergogne-Berezin, E. 2001. The increasing role of Acinetobacter species as nosocomial pathogens. Curr. Infect. Dis. Rep. 3440-444. - PubMed
-
- Bouvet, P. J. M., and P. A. D. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36228-240.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

