Functional dissection of the human TNRC6 (GW182-related) family of proteins
- PMID: 19470757
- PMCID: PMC2715800
- DOI: 10.1128/MCB.00380-09
Functional dissection of the human TNRC6 (GW182-related) family of proteins
Abstract
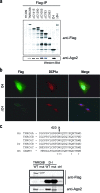
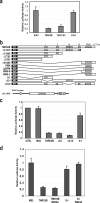
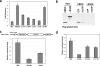
Argonaute (Ago) proteins through their association with small RNAs perform a critical function in the effector step of RNA interference. The TNRC6 (trinucleotide repeat containing 6) family of proteins have been shown to stably associate with Agos in mammalian cells. Here, we describe the isolation and functional characterization of TNRC6B- and TNRC6C-containing complexes. We show that TNRC6B and TNRC6C proteins associate with all four human Agos which are already loaded with microRNAs. Detailed domain analysis of TNRC6B protein indicated that distinct domains of the protein are required for Ago binding and P-body localization. Functional analysis using reporter constructs responsive to TNRC6B tethered through an MS2-binding domain indicates that neither the Ago-binding nor the P-body localization domains are required for translational silencing. In contrast, the C-terminal domain containing the RNA recognition motif plays a critical role in the silencing mediated by the TNRC6B protein.
Figures




References
-
- Bagga, S., J. Bracht, S. Hunter, K. Massirer, J. Holtz, R. Eachus, and A. E. Pasquinelli. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122553-563. - PubMed
-
- Behlke, M. A., S. A. Dames, W. H. McDonald, K. L. Gould, E. J. Devor, and J. A. Walder. 2000. Use of high specific activity StarFire oligonucleotide probes to visualize low-abundance pre-mRNA splicing intermediates in S. pombe. BioTechniques 29892-897. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
