Lack of association between polymorphisms of the IL18R1 and IL18RAP genes and cardiovascular risk: the MORGAM Project
- PMID: 19473509
- PMCID: PMC2692850
- DOI: 10.1186/1471-2350-10-44
Lack of association between polymorphisms of the IL18R1 and IL18RAP genes and cardiovascular risk: the MORGAM Project
Abstract
Background: Interleukin-18 is a pro-inflammatory cytokine suspected to be associated with atherosclerosis and its complications. We had previously shown that one single nucleotide polymorphism (SNP) of the IL18 gene was associated with cardiovascular disease (CVD) through an interaction with smoking. As a further step for elucidating the contribution of the IL-18 pathway to the etiology of CVD, we here investigated the association between the genetic variability of two IL-18 receptor genes, IL18R1 and IL18RAP, with the risk of developing CVD.
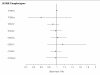
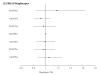
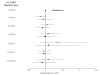
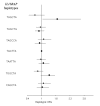
Methods: Eleven tagging SNPs, 5 in IL18R1 and 6 in IL18RAP, characterizing the haplotypic variability of the corresponding genes; were genotyped in 5 European prospective CVD cohorts including 1416 cases and 1772 non-cases, as part of the MORGAM project. Both single-locus and haplotypes analyses were carried out to investigate the association of these SNPs with CVD.
Results: We did not find any significant differences in allele, genotype and haplotype frequencies between cases and non-cases for either of the two genes. Moreover, the search for interactions between SNPs located in different genes, including 5 IL18 SNPs previously studied in the MORGAM project, and between SNPs and environmental factors remained unfruitful.
Conclusion: Our analysis suggests that the variability of IL18R1 and IL18RAP genes are unlikely to contribute to modulate the risk of CVD.
Figures




Similar articles
-
Polymorphisms of ST2-IL18R1-IL18RAP gene cluster: a new risk for autoimmune thyroid diseases.Int J Immunogenet. 2016 Feb;43(1):18-24. doi: 10.1111/iji.12240. Epub 2015 Nov 14. Int J Immunogenet. 2016. PMID: 26566691
-
Haplotypic analysis of tag SNPs of the interleukin-18 gene in relation to cardiovascular disease events: the MORGAM Project.Eur J Hum Genet. 2008 Dec;16(12):1512-20. doi: 10.1038/ejhg.2008.127. Epub 2008 Jul 16. Eur J Hum Genet. 2008. PMID: 18628791
-
Association of IL1RL1, IL18R1, and IL18RAP gene cluster polymorphisms with asthma and atopy.J Allergy Clin Immunol. 2008 Sep;122(3):651-4.e8. doi: 10.1016/j.jaci.2008.06.030. J Allergy Clin Immunol. 2008. PMID: 18774397 No abstract available.
-
IL18-family Genes Polymorphism Is Associated with the Risk of Myocardial Infarction and IL18 Concentration in Patients with Coronary Artery Disease.Immunol Invest. 2022 May;51(4):802-816. doi: 10.1080/08820139.2021.1876085. Epub 2021 Feb 1. Immunol Invest. 2022. PMID: 33522333
-
Common SNPs/haplotypes in IL18R1 and IL18 genes are associated with variations in humoral immunity to smallpox vaccination in Caucasians and African Americans.J Infect Dis. 2011 Aug 1;204(3):433-41. doi: 10.1093/infdis/jir268. J Infect Dis. 2011. PMID: 21742843 Free PMC article.
Cited by
-
The role of interleukin-18 in the metabolic syndrome.Cardiovasc Diabetol. 2010 Mar 23;9:11. doi: 10.1186/1475-2840-9-11. Cardiovasc Diabetol. 2010. PMID: 20331890 Free PMC article. Review.
-
Molecular Characterization of the NLRC4 Expression in Relation to Interleukin-18 Levels.Circ Cardiovasc Genet. 2015 Oct;8(5):717-26. doi: 10.1161/CIRCGENETICS.115.001079. Epub 2015 Sep 11. Circ Cardiovasc Genet. 2015. PMID: 26362438 Free PMC article.
-
Transcriptome profiles of the skeletal muscle of mature cows during feed restriction and realimentation.BMC Res Notes. 2021 Sep 16;14(1):361. doi: 10.1186/s13104-021-05757-8. BMC Res Notes. 2021. PMID: 34530907 Free PMC article.
-
Canonical correlation analysis for gene-based pleiotropy discovery.PLoS Comput Biol. 2014 Oct 16;10(10):e1003876. doi: 10.1371/journal.pcbi.1003876. eCollection 2014 Oct. PLoS Comput Biol. 2014. PMID: 25329069 Free PMC article.
-
Identification of candidate biomarkers and therapeutic agents for heart failure by bioinformatics analysis.BMC Cardiovasc Disord. 2021 Jul 4;21(1):329. doi: 10.1186/s12872-021-02146-8. BMC Cardiovasc Disord. 2021. PMID: 34218797 Free PMC article.
References
-
- de Nooijer R, Thusen JH von der, Verkleij CJ, Kuiper J, Jukema JW, Wall EE van der, van Berkel JC, Biessen EA. Overexpression of IL-18 decreases intimal collagen content and promotes a vulnerable plaque phenotype in apolipoprotein-E-deficient mice. Arterioscler Thromb Vasc Biol. 2004;24:2313–2319. doi: 10.1161/01.ATV.0000147126.99529.0a. - DOI - PubMed
-
- Mallat Z, Heymes C, Corbaz A, Logeart D, Alouani S, Cohen-Solal A, Seidler T, Hasenfuss G, Chvatchko Y, Shah AM, Tedgui A. Evidence for altered interleukin 18 (IL)-18 pathway in human heart failure. Faseb J. 2004;18:1752–1754. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

