Hypermethylation of genomic 3.3-kb repeats is frequent event in HPV-positive cervical cancer
- PMID: 19473516
- PMCID: PMC2695481
- DOI: 10.1186/1755-8794-2-30
Hypermethylation of genomic 3.3-kb repeats is frequent event in HPV-positive cervical cancer
Abstract
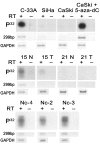
Background: Large-scale screening methods are widely used to reveal cancer-specific DNA methylation markers. We previously identified non-satellite 3.3-kb repeats associated with facioscapulohumeral muscular dystrophy (FSHD) as hypermethylated in cervical cancer in genome-wide screening. To determine whether hypermethylation of 3.3-kb repeats is a tumor-specific event and to evaluate frequency of this event in tumors, we investigated the 3.3-kb repeat methylation status in human papilloma virus (HPV)-positive cervical tumors, cancer cell lines, and normal cervical tissues. Open reading frames encoding DUX family proteins are contained within some 3.3-kb repeat units. The DUX mRNA expression profile was also studied in these tissues.
Methods: The methylation status of 3.3-kb repeats was evaluated by Southern blot hybridization and bisulfite genomic sequencing. The expression of DUX mRNA was analyzed by RT-PCR and specificity of PCR products was confirmed by sequencing analysis.
Results: Hypermethylation of 3.3-kb repeats relative to normal tissues was revealed for the first time in more than 50% (18/34) of cervical tumors and in 4 HPV-positive cervical cancer cell lines. Hypermethylation of 3.3-kb repeats was observed in tumors concurrently with or independently of hypomethylation of classical satellite 2 sequences (Sat2) that were hypomethylated in 75% (15/20) of cervical tumors. We have revealed the presence of transcripts highly homologous to DUX4 and DUX10 genes in normal tissues and down-regulation of transcripts in 68% of tumors with and without 3.3-kb repeats hypermethylation.
Conclusion: Our results demonstrate that hypermethylation rather than hypomethylation of 3.3-kb repeats is the predominant event in HPV-associated cervical cancer and provide new insight into the epigenetic changes of repetitive DNA elements in carcinogenesis.
Figures



Similar articles
-
Up-regulation of expression and lack of 5' CpG island hypermethylation of p16 INK4a in HPV-positive cervical carcinomas.BMC Cancer. 2007 Mar 14;7:47. doi: 10.1186/1471-2407-7-47. BMC Cancer. 2007. PMID: 17359536 Free PMC article.
-
Identifying diagnostic DNA methylation profiles for facioscapulohumeral muscular dystrophy in blood and saliva using bisulfite sequencing.Clin Epigenetics. 2014 Oct 29;6(1):23. doi: 10.1186/1868-7083-6-23. eCollection 2014. Clin Epigenetics. 2014. PMID: 25400706 Free PMC article.
-
Methylation of the FSHD syndrome-linked subtelomeric repeat in normal and FSHD cell cultures and tissues.Mol Genet Metab. 2001 Nov;74(3):322-31. doi: 10.1006/mgme.2001.3219. Mol Genet Metab. 2001. PMID: 11708861
-
Cancer-linked DNA hypomethylation and its relationship to hypermethylation.Curr Top Microbiol Immunol. 2006;310:251-74. doi: 10.1007/3-540-31181-5_12. Curr Top Microbiol Immunol. 2006. PMID: 16909914 Review.
-
DNA hypomethylation is prevalent even in low-grade breast cancers.Cancer Biol Ther. 2004 Dec;3(12):1225-31. doi: 10.4161/cbt.3.12.1222. Epub 2004 Dec 9. Cancer Biol Ther. 2004. PMID: 15539937 Review.
Cited by
-
Epigenetic reprogramming of host genes in viral and microbial pathogenesis.Trends Microbiol. 2010 Oct;18(10):439-47. doi: 10.1016/j.tim.2010.07.003. Epub 2010 Aug 18. Trends Microbiol. 2010. PMID: 20724161 Free PMC article. Review.
-
Cancer-related genes in the transcription signature of facioscapulohumeral dystrophy myoblasts and myotubes.J Cell Mol Med. 2014 Feb;18(2):208-17. doi: 10.1111/jcmm.12182. Epub 2013 Dec 17. J Cell Mol Med. 2014. PMID: 24341522 Free PMC article.
-
Diagnostic approach for FSHD revisited: SMCHD1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1.Eur J Hum Genet. 2015 Jun;23(6):808-16. doi: 10.1038/ejhg.2014.191. Epub 2014 Nov 5. Eur J Hum Genet. 2015. PMID: 25370034 Free PMC article.
-
DNA hypomethylation in cancer cells.Epigenomics. 2009 Dec;1(2):239-59. doi: 10.2217/epi.09.33. Epigenomics. 2009. PMID: 20495664 Free PMC article. Review.
-
[DNA methylation. From basic research to routine diagnostics].Pathologe. 2010 Oct;31 Suppl 2:274-9. doi: 10.1007/s00292-010-1300-7. Pathologe. 2010. PMID: 20824434 German.
References
-
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. - PubMed
LinkOut - more resources
Full Text Sources

