Stabilization of the E* form turns thrombin into an anticoagulant
- PMID: 19473969
- PMCID: PMC2740429
- DOI: 10.1074/jbc.M109.012344
Stabilization of the E* form turns thrombin into an anticoagulant
Abstract
Previous studies have shown that deletion of nine residues in the autolysis loop of thrombin produces a mutant with an anticoagulant propensity of potential clinical relevance, but the molecular origin of the effect has remained unresolved. The x-ray crystal structure of this mutant solved in the free form at 1.55 A resolution reveals an inactive conformation that is practically identical (root mean square deviation of 0.154 A) to the recently identified E* form. The side chain of Trp(215) collapses into the active site by shifting > 10 A from its position in the active E form, and the oxyanion hole is disrupted by a flip of the Glu(192)-Gly(193) peptide bond. This finding confirms the existence of the inactive form E* in essentially the same incarnation as first identified in the structure of the thrombin mutant D102N. In addition, it demonstrates that the anticoagulant profile often caused by a mutation of the thrombin scaffold finds its likely molecular origin in the stabilization of the inactive E* form that is selectively shifted to the active E form upon thrombomodulin and protein C binding.
Figures



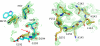
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

