Identification of the C1q-binding Sites of Human C1r and C1s: a refined three-dimensional model of the C1 complex of complement
- PMID: 19473974
- PMCID: PMC2740559
- DOI: 10.1074/jbc.M109.004473
Identification of the C1q-binding Sites of Human C1r and C1s: a refined three-dimensional model of the C1 complex of complement
Abstract
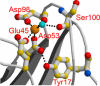
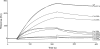
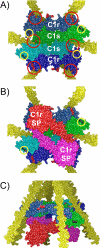
The C1 complex of complement is assembled from a recognition protein C1q and C1s-C1r-C1r-C1s, a Ca(2+)-dependent tetramer of two modular proteases C1r and C1s. Resolution of the x-ray structure of the N-terminal CUB(1)-epidermal growth factor (EGF) C1s segment has led to a model of the C1q/C1s-C1r-C1r-C1s interaction where the C1q collagen stem binds at the C1r/C1s interface through ionic bonds involving acidic residues contributed by the C1r EGF module (Gregory, L. A., Thielens, N. M., Arlaud, G. J., Fontecilla-Camps, J. C., and Gaboriaud, C. (2003) J. Biol. Chem. 278, 32157-32164). To identify the C1q-binding sites of C1s-C1r-C1r-C1s, a series of C1r and C1s mutants was expressed, and the C1q binding ability of the resulting tetramer variants was assessed by surface plasmon resonance. Mutations targeting the Glu(137)-Glu-Asp(139) stretch in the C1r EGF module had no effect on C1 assembly, ruling out our previous interaction model. Additional mutations targeting residues expected to participate in the Ca(2+)-binding sites of the C1r and C1s CUB modules provided evidence for high affinity C1q-binding sites contributed by the C1r CUB(1) and CUB(2) modules and lower affinity sites contributed by C1s CUB(1). All of the sites implicate acidic residues also contributing Ca(2+) ligands. C1s-C1r-C1r-C1s thus contributes six C1q-binding sites, one per C1q stem. Based on the location of these sites and available structural information, we propose a refined model of C1 assembly where the CUB(1)-EGF-CUB(2) interaction domains of C1r and C1s are entirely clustered inside C1q and interact through six binding sites with reactive lysines of the C1q stems. This mechanism is similar to that demonstrated for mannan-binding lectin (MBL)-MBL-associated serine protease and ficolin-MBL-associated serine protease complexes.
Figures

References
-
- Cooper N. R. (1985) Adv. Immunol. 37, 151–216 - PubMed
-
- Arlaud G. J., Colomb M. G., Gagnon J. (1987) Immunol. Today 8, 106–111 - PubMed
-
- Arlaud G. J., Gaboriaud C., Thielens N. M., Rossi V., Bersch B., Hernandez J. F., Fontecilla-Camps J. C. (2001) Immunol. Rev. 180, 136–145 - PubMed
-
- Kishore U., Reid K. B. M. (2000) Immunopharmacology 49, 159–170 - PubMed
-
- Bork P., Beckmann G. (1993) J. Mol. Biol. 231, 539–545 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

