Repeated domains of leptospira immunoglobulin-like proteins interact with elastin and tropoelastin
- PMID: 19473986
- PMCID: PMC2740563
- DOI: 10.1074/jbc.M109.004531
Repeated domains of leptospira immunoglobulin-like proteins interact with elastin and tropoelastin
Abstract
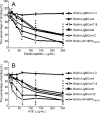
Leptospira spp., the causative agents of leptospirosis, adhere to components of the extracellular matrix, a pivotal role for colonization of host tissues during infection. Previously, we and others have shown that Leptospira immunoglobulin-like proteins (Lig) of Leptospira spp. bind to fibronectin, laminin, collagen, and fibrinogen. In this study, we report that Leptospira can be immobilized by human tropoelastin (HTE) or elastin from different tissues, including lung, skin, and blood vessels, and that Lig proteins can bind to HTE or elastin. Moreover, both elastin and HTE bind to the same LigB immunoglobulin-like domains, including LigBCon4, LigBCen7'-8, LigBCen9, and LigBCen12 as demonstrated by enzyme-linked immunosorbent assay (ELISA) and competition ELISAs. The LigB immunoglobulin-like domain binds to the 17th to 27th exons of HTE (17-27HTE) as determined by ELISA (LigBCon4, K(D) = 0.50 microm; LigBCen7'-8, K(D) = 0.82 microm; LigBCen9, K(D) = 1.54 microm; and LigBCen12, K(D) = 0.73 microm). The interaction of LigBCon4 and 17-27HTE was further confirmed by steady state fluorescence spectroscopy (K(D) = 0.49 microm) and ITC (K(D) = 0.54 microm). Furthermore, the binding was enthalpy-driven and affected by environmental pH, indicating it is a charge-charge interaction. The binding affinity of LigBCon4D341N to 17-27HTE was 4.6-fold less than that of wild type LigBCon4. In summary, we show that Lig proteins of Leptospira spp. interact with elastin and HTE, and we conclude this interaction may contribute to Leptospira adhesion to host tissues during infection.
Figures




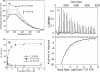
 ). The thermodynamic parameters are shown in Table 3.
). The thermodynamic parameters are shown in Table 3.

 ). The thermodynamic parameters are shown in Table 3.
). The thermodynamic parameters are shown in Table 3.Similar articles
-
The terminal immunoglobulin-like repeats of LigA and LigB of Leptospira enhance their binding to gelatin binding domain of fibronectin and host cells.PLoS One. 2010 Jun 24;5(6):e11301. doi: 10.1371/journal.pone.0011301. PLoS One. 2010. PMID: 20585579 Free PMC article.
-
Leptospira Immunoglobulin-Like Protein B Interacts with the 20th Exon of Human Tropoelastin Contributing to Leptospiral Adhesion to Human Lung Cells.Front Cell Infect Microbiol. 2017 May 9;7:163. doi: 10.3389/fcimb.2017.00163. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28536676 Free PMC article.
-
The OmpL37 surface-exposed protein is expressed by pathogenic Leptospira during infection and binds skin and vascular elastin.PLoS Negl Trop Dis. 2010 Sep 7;4(9):e815. doi: 10.1371/journal.pntd.0000815. PLoS Negl Trop Dis. 2010. PMID: 20844573 Free PMC article.
-
Leptospiral Immunoglobulin-Like Domain Proteins: Roles in Virulence and Immunity.Front Immunol. 2021 Jan 8;11:579907. doi: 10.3389/fimmu.2020.579907. eCollection 2020. Front Immunol. 2021. PMID: 33488581 Free PMC article. Review.
-
Elastin: relation of protein and gene structure to disease.Lab Invest. 1984 Dec;51(6):605-23. Lab Invest. 1984. PMID: 6150137 Review.
Cited by
-
Pathogenic Leptospira species acquire factor H and vitronectin via the surface protein LcpA.Infect Immun. 2015 Mar;83(3):888-97. doi: 10.1128/IAI.02844-14. Epub 2014 Dec 22. Infect Immun. 2015. PMID: 25534939 Free PMC article.
-
Staphylococcus aureus manganese transport protein C (MntC) is an extracellular matrix- and plasminogen-binding protein.PLoS One. 2014 Nov 19;9(11):e112730. doi: 10.1371/journal.pone.0112730. eCollection 2014. PLoS One. 2014. PMID: 25409527 Free PMC article.
-
Identification of lysine residues in the Borrelia burgdorferi DbpA adhesin required for murine infection.Infect Immun. 2014 Aug;82(8):3186-98. doi: 10.1128/IAI.02036-14. Epub 2014 May 19. Infect Immun. 2014. PMID: 24842928 Free PMC article.
-
Strain-specific variation of the decorin-binding adhesin DbpA influences the tissue tropism of the lyme disease spirochete.PLoS Pathog. 2014 Jul 31;10(7):e1004238. doi: 10.1371/journal.ppat.1004238. eCollection 2014 Jul. PLoS Pathog. 2014. PMID: 25079227 Free PMC article.
-
Leptospira: a spirochaete with a hybrid outer membrane.Mol Microbiol. 2010 Aug;77(4):805-14. doi: 10.1111/j.1365-2958.2010.07262.x. Epub 2010 Jun 28. Mol Microbiol. 2010. PMID: 20598085 Free PMC article. Review.
References
-
- Palaniappan R. U., Ramanujam S., Chang Y. F. (2007) Curr. Opin. Infect. Dis. 20, 284–292 - PubMed
-
- Faine S. B., Adher B., Bolin C., Perolat P. (eds) (1999) Leptospira and Leptospirosis., pp. 67–69, MedSci, Medbourne, Australia
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

