Towards a quantitative understanding of the within-host dynamics of influenza A infections
- PMID: 19474085
- PMCID: PMC2839376
- DOI: 10.1098/rsif.2009.0067
Towards a quantitative understanding of the within-host dynamics of influenza A infections
Abstract
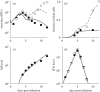
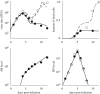
Although the influenza A virus has been extensively studied, a quantitative understanding of the infection dynamics is still lacking. To make progress in this direction, we designed several mathematical models and compared them with data from influenza A infections of mice. We find that the immune response (IR) plays an important part in the infection dynamics. Both an innate and an adaptive IR are required to provide adequate explanation of the data. In contrast, regrowth of epithelial cells did not seem to be an important mechanism on the time scale of the infection. We also find that different model variants for both innate and adaptive responses fit the data well, indicating the need for additional data to allow further model discrimination.
Figures
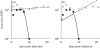
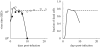
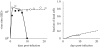
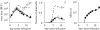
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
