Human bocavirus can be cultured in differentiated human airway epithelial cells
- PMID: 19474096
- PMCID: PMC2708629
- DOI: 10.1128/JVI.00614-09
Human bocavirus can be cultured in differentiated human airway epithelial cells
Abstract
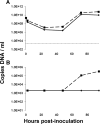
In 2005, a human bocavirus was discovered in children with respiratory tract illnesses. Attempts to culture this virus on conventional cell lines has failed thus far. We investigated whether the virus can replicate on pseudostratified human airway epithelium. This cell culture system mimics the human airway environment and facilitates culturing of various respiratory agents. The cells were inoculated with human bocavirus-positive nasopharyngeal washes from children, and virus replication was monitored by measuring apical release of the virus via real-time PCR. Furthermore, we identified different viral mRNAs in the infected cells. All mRNAs were transcribed from a single promoter but varied due to alternative splicing and alternative polyadenylation, similar to what has been described for bovine parvovirus and minute virus of canines, the other two members of the Bocavirus genus. Thus, transcription of human bocavirus displays strong homology to the transcription of the other bocaviruses. In conclusion, we report here for the first time that human bocavirus can be propagated in an in vitro culture system and present a detailed map of the set of mRNAs that are produced by the virus.
Figures





References
-
- Berns, K., and C. R. Parrish. 2007. Parvoviridae, p. 2437-2477. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

