Mutagenesis of varicella-zoster virus glycoprotein B: putative fusion loop residues are essential for viral replication, and the furin cleavage motif contributes to pathogenesis in skin tissue in vivo
- PMID: 19474103
- PMCID: PMC2708640
- DOI: 10.1128/JVI.00400-09
Mutagenesis of varicella-zoster virus glycoprotein B: putative fusion loop residues are essential for viral replication, and the furin cleavage motif contributes to pathogenesis in skin tissue in vivo
Abstract
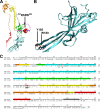
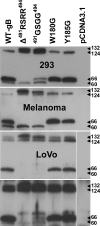
Glycoprotein B (gB), the most conserved protein in the family Herpesviridae, is essential for the fusion of viral and cellular membranes. Information about varicella-zoster virus (VZV) gB is limited, but homology modeling showed that the structure of VZV gB was similar to that of herpes simplex virus (HSV) gB, including the putative fusion loops. In contrast to HSV gB, VZV gB had a furin recognition motif ([R]-X-[KR]-R-|-X, where | indicates the position at which the polypeptide is cleaved) at residues 491 to 494, thought to be required for gB cleavage into two polypeptides. To investigate their contribution, the putative primary fusion loop or the furin recognition motif was mutated in expression constructs and in the context of the VZV genome. Substitutions in the primary loop, W180G and Y185G, plus the deletion mutation Delta491RSRR494 and point mutation 491GSGG494 in the furin recognition motif did not affect gB expression or cellular localization in transfected cells. Infectious VZV was recovered from parental Oka (pOka)-bacterial artificial chromosomes that had either the Delta491RSRR494 or 491GSGG494 mutation but not the point mutations W180G and Y185G, demonstrating that residues in the primary loop of gB were essential but gB cleavage was not required for VZV replication in vitro. Virion morphology, protein localization, plaque size, and replication were unaffected for the pOka-gBDelta491RSRR494 or pOka-gB491GSGG494 virus compared to pOka in vitro. However, deletion of the furin recognition motif caused attenuation of VZV replication in human skin xenografts in vivo. This is the first evidence that cleavage of a herpesvirus fusion protein contributes to viral pathogenesis in vivo, as seen for fusion proteins in other virus families.
Figures




Similar articles
-
Varicella-zoster virus: molecular controls of cell fusion-dependent pathogenesis.Biochem Soc Trans. 2020 Dec 18;48(6):2415-2435. doi: 10.1042/BST20190511. Biochem Soc Trans. 2020. PMID: 33259590 Free PMC article. Review.
-
The Glycoprotein B Cytoplasmic Domain Lysine Cluster Is Critical for Varicella-Zoster Virus Cell-Cell Fusion Regulation and Infection.J Virol. 2016 Dec 16;91(1):e01707-16. doi: 10.1128/JVI.01707-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795427 Free PMC article.
-
Regulation of the ORF61 promoter and ORF61 functions in varicella-zoster virus replication and pathogenesis.J Virol. 2009 Aug;83(15):7560-72. doi: 10.1128/JVI.00118-09. Epub 2009 May 20. J Virol. 2009. PMID: 19457996 Free PMC article.
-
The immediate-early 63 protein of Varicella-Zoster virus: analysis of functional domains required for replication in vitro and for T-cell and skin tropism in the SCIDhu model in vivo.J Virol. 2004 Feb;78(3):1181-94. doi: 10.1128/jvi.78.3.1181-1194.2004. J Virol. 2004. PMID: 14722273 Free PMC article.
-
Investigation of varicella-zoster virus neurotropism and neurovirulence using SCID mouse-human DRG xenografts.J Neurovirol. 2011 Dec;17(6):570-7. doi: 10.1007/s13365-011-0066-x. Epub 2011 Dec 8. J Neurovirol. 2011. PMID: 22161683 Review.
Cited by
-
Varicella-zoster virus: molecular controls of cell fusion-dependent pathogenesis.Biochem Soc Trans. 2020 Dec 18;48(6):2415-2435. doi: 10.1042/BST20190511. Biochem Soc Trans. 2020. PMID: 33259590 Free PMC article. Review.
-
Fatal Elephant Endotheliotropic Herpesvirus Infection of Two Young Asian Elephants.Microorganisms. 2019 Sep 26;7(10):396. doi: 10.3390/microorganisms7100396. Microorganisms. 2019. PMID: 31561506 Free PMC article.
-
Cellular transcription factor YY1 mediates the varicella-zoster virus (VZV) IE62 transcriptional activation.Virology. 2014 Jan 20;449:244-53. doi: 10.1016/j.virol.2013.11.029. Epub 2013 Dec 12. Virology. 2014. PMID: 24418559 Free PMC article.
-
ORF7 of Varicella-Zoster Virus Is Required for Viral Cytoplasmic Envelopment in Differentiated Neuronal Cells.J Virol. 2017 May 26;91(12):e00127-17. doi: 10.1128/JVI.00127-17. Print 2017 Jun 15. J Virol. 2017. PMID: 28356523 Free PMC article.
-
Nectin-2 Acts as a Viral Entry Mediated Molecule That Binds to Human Herpesvirus 6B Glycoprotein B.Viruses. 2022 Jan 16;14(1):160. doi: 10.3390/v14010160. Viruses. 2022. PMID: 35062364 Free PMC article.
References
-
- Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 1136-42.
-
- Arbeter, A. M., S. E. Starr, R. E. Weibel, and S. A. Plotkin. 1982. Live attenuated varicella vaccine: immunization of healthy children with the OKA strain. J. Pediatr. 100886-893. - PubMed
-
- Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22195-201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

