Processing bodies are not required for mammalian nonsense-mediated mRNA decay
- PMID: 19474145
- PMCID: PMC2704072
- DOI: 10.1261/rna.1672509
Processing bodies are not required for mammalian nonsense-mediated mRNA decay
Abstract
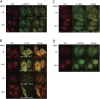
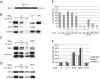
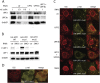
Nonsense-mediated mRNA decay (NMD) is a eukaryotic quality-control mechanism that recognizes and degrades mRNAs with premature termination codons (PTCs). In yeast, PTC-containing mRNAs are targeted to processing bodies (P-bodies), and yeast strains expressing an ATPase defective Upf1p mutant accumulate P-bodies. Here we show that in human cells, an ATPase-deficient UPF1 mutant and a fraction of UPF2 and UPF3b accumulate in cytoplasmic foci that co-localize with P-bodies. Depletion of the P-body component Ge-1, which prevents formation of microscopically detectable P-bodies, also impairs the localization of mutant UPF1, UPF2, and UPF3b in cytoplasmic foci. However, the accumulation of the ATPase-deficient UPF1 mutant in P-bodies is independent of UPF2, UPF3b, or SMG1, and the ATPase-deficient UPF1 mutant can localize into the P-bodies independent of its phosphorylation status. Most importantly, disruption of P-bodies by depletion of Ge-1 affects neither the mRNA levels of PTC-containing reporter genes nor endogenous NMD substrates. Consistent with the recently reported decapping-independent SMG6-mediated endonucleolytic decay of human nonsense mRNAs, our results imply that microscopically detectable P-bodies are not required for mammalian NMD.
Figures

Similar articles
-
Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways.RNA. 2013 Oct;19(10):1432-48. doi: 10.1261/rna.038893.113. Epub 2013 Aug 20. RNA. 2013. PMID: 23962664 Free PMC article.
-
CK2-mediated TEL2 phosphorylation augments nonsense-mediated mRNA decay (NMD) by increase of SMG1 stability.Biochim Biophys Acta. 2013 Oct;1829(10):1047-55. doi: 10.1016/j.bbagrm.2013.06.002. Epub 2013 Jul 3. Biochim Biophys Acta. 2013. PMID: 23831331
-
UPF1 P-body localization.Biochem Soc Trans. 2008 Aug;36(Pt 4):698-700. doi: 10.1042/BST0360698. Biochem Soc Trans. 2008. PMID: 18631143 Review.
-
Targeting of aberrant mRNAs to cytoplasmic processing bodies.Cell. 2006 Jun 16;125(6):1095-109. doi: 10.1016/j.cell.2006.04.037. Cell. 2006. PMID: 16777600 Free PMC article.
-
Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay.Genes Cells. 2013 Mar;18(3):161-75. doi: 10.1111/gtc.12033. Epub 2013 Jan 28. Genes Cells. 2013. PMID: 23356578 Review.
Cited by
-
Ribosome dynamics and mRNA turnover, a complex relationship under constant cellular scrutiny.Wiley Interdiscip Rev RNA. 2021 Nov;12(6):e1658. doi: 10.1002/wrna.1658. Epub 2021 May 5. Wiley Interdiscip Rev RNA. 2021. PMID: 33949788 Free PMC article. Review.
-
Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors.Cell Mol Life Sci. 2010 Mar;67(5):677-700. doi: 10.1007/s00018-009-0177-1. Epub 2009 Oct 27. Cell Mol Life Sci. 2010. PMID: 19859661 Free PMC article. Review.
-
Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis.Mol Cancer Res. 2010 Mar;8(3):295-308. doi: 10.1158/1541-7786.MCR-09-0502. Epub 2010 Feb 23. Mol Cancer Res. 2010. PMID: 20179151 Free PMC article. Review.
-
Chromatoid Body Protein TDRD6 Supports Long 3' UTR Triggered Nonsense Mediated mRNA Decay.PLoS Genet. 2016 May 5;12(5):e1005857. doi: 10.1371/journal.pgen.1005857. eCollection 2016 May. PLoS Genet. 2016. PMID: 27149095 Free PMC article.
-
LSM14B controls oocyte mRNA storage and stability to ensure female fertility.Cell Mol Life Sci. 2023 Aug 14;80(9):247. doi: 10.1007/s00018-023-04898-2. Cell Mol Life Sci. 2023. PMID: 37578641 Free PMC article.
References
-
- Behm-Ansmant I, Izaurralde E. Quality control of gene expression: A stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes & Dev. 2006;20:391–398. - PubMed
-
- Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: An ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. - PubMed
-
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
