UBXD4, a UBX-containing protein, regulates the cell surface number and stability of alpha3-containing nicotinic acetylcholine receptors
- PMID: 19474315
- PMCID: PMC2935801
- DOI: 10.1523/JNEUROSCI.4723-08.2009
UBXD4, a UBX-containing protein, regulates the cell surface number and stability of alpha3-containing nicotinic acetylcholine receptors
Abstract
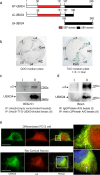
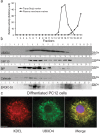
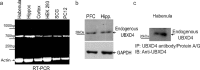
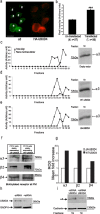
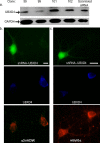
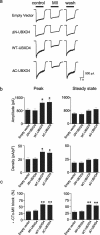
Adaptor proteins are likely to modulate spatially and temporally the trafficking of a number of membrane proteins, including neuronal nicotinic acetylcholine receptors (nAChRs). A yeast two-hybrid screen identified a novel UBX-containing protein, UBXD4, as one of the cytosolic proteins that interact directly with the alpha3 and alpha4 nAChR subunits. The function of UBX-containing proteins is largely unknown. Immunoprecipitation and confocal microscopy confirmed the interaction of UBXD4 with alpha3-containing nAChRs (alpha3* nAChRs) expressed in HEK293 cells, PC12 cells, and rat cortical neurons. Overexpression of UBXD4 in differentiated PC12 cells (dPC12) increased nAChR cell surface expression, especially that of the alpha3beta2 subtype. These findings were corroborated by electrophysiology, immunofluorescent staining, and biotinylation of surface receptors. Silencing of UBXD4 led to a significant reduction of alpha3* nAChRs in rat cortical neurons and dPC12 cells. Biochemical and immunofluorescence studies of endogenous UBXD4 showed that the protein is located in both the ER and cis-Golgi compartments. Our investigations also showed that the alpha3 subunit is ubiquitinated and that UBXD4 can interfere with its ubiquitination and consequent degradation by the proteasome. Our data suggest that UBXD4 modulates the distribution of alpha3* nAChRs between specialized intracellular compartments and the plasma membrane. This effect is achieved by controlling the stability of the alpha3 subunit and, consequently, the number of receptors at the cell surface.
Figures



Similar articles
-
The ubiquitin-proteasome system regulates the stability of neuronal nicotinic acetylcholine receptors.J Mol Neurosci. 2010 Jan;40(1-2):177-84. doi: 10.1007/s12031-009-9272-x. Epub 2009 Aug 20. J Mol Neurosci. 2010. PMID: 19693707 Free PMC article.
-
UBXN2A regulates nicotinic receptor degradation by modulating the E3 ligase activity of CHIP.Biochem Pharmacol. 2015 Oct 15;97(4):518-530. doi: 10.1016/j.bcp.2015.08.084. Epub 2015 Aug 8. Biochem Pharmacol. 2015. PMID: 26265139 Free PMC article.
-
Engineering neuronal nicotinic acetylcholine receptors with functional sensitivity to alpha-bungarotoxin: a novel alpha3-knock-in mouse.Eur J Neurosci. 2009 Dec 3;30(11):2064-76. doi: 10.1111/j.1460-9568.2009.07016.x. Epub 2009 Nov 25. Eur J Neurosci. 2009. PMID: 20128845 Free PMC article.
-
A novel nicotinic acetylcholine receptor subtype in basal forebrain cholinergic neurons with high sensitivity to amyloid peptides.J Neurosci. 2009 Jan 28;29(4):918-29. doi: 10.1523/JNEUROSCI.3952-08.2009. J Neurosci. 2009. PMID: 19176801 Free PMC article.
-
Ric-3 promotes alpha7 nicotinic receptor assembly and trafficking through the ER subcompartment of dendrites.J Neurosci. 2010 Jul 28;30(30):10112-26. doi: 10.1523/JNEUROSCI.6344-09.2010. J Neurosci. 2010. PMID: 20668195 Free PMC article.
Cited by
-
The ubiquitin-proteasome system regulates the stability of neuronal nicotinic acetylcholine receptors.J Mol Neurosci. 2010 Jan;40(1-2):177-84. doi: 10.1007/s12031-009-9272-x. Epub 2009 Aug 20. J Mol Neurosci. 2010. PMID: 19693707 Free PMC article.
-
Nicotine-induced upregulation of native neuronal nicotinic receptors is caused by multiple mechanisms.J Neurosci. 2012 Feb 8;32(6):2227-38. doi: 10.1523/JNEUROSCI.5438-11.2012. J Neurosci. 2012. PMID: 22323734 Free PMC article.
-
Elucidation of molecular impediments in the α6 subunit for in vitro expression of functional α6β4* nicotinic acetylcholine receptors.J Biol Chem. 2013 Nov 22;288(47):33708-33721. doi: 10.1074/jbc.M113.509356. Epub 2013 Oct 1. J Biol Chem. 2013. PMID: 24085295 Free PMC article.
-
Nucleocytoplasmic Translocation of UBXN2A Is Required for Apoptosis during DNA Damage Stresses in Colon Cancer Cells.J Cancer. 2015 Sep 3;6(11):1066-78. doi: 10.7150/jca.12134. eCollection 2015. J Cancer. 2015. PMID: 26516353 Free PMC article.
-
Nicotine-modulated subunit stoichiometry affects stability and trafficking of α3β4 nicotinic receptor.J Neurosci. 2013 Jul 24;33(30):12316-28. doi: 10.1523/JNEUROSCI.2393-13.2013. J Neurosci. 2013. PMID: 23884938 Free PMC article.
References
-
- Arias HR, Bhumireddy P. Anesthetics as chemical tools to study the structure and function of nicotinic acetylcholine receptors. Curr Protein Pept Sci. 2005;6:451–472. - PubMed
-
- Bedford FK, Kittler JT, Muller E, Thomas P, Uren JM, Merlo D, Wisden W, Triller A, Smart TG, Moss SJ. GABA(A) receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat Neurosci. 2001;4:908–916. - PubMed
-
- Belizario JE, Alves J, Garay-Malpartida M, Occhiucci JM. Coupling caspase cleavage and proteasomal degradation of proteins carrying PEST motif. Curr Protein Pept Sci. 2008;9:210–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
