Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila
- PMID: 19474317
- PMCID: PMC6665603
- DOI: 10.1523/JNEUROSCI.1585-09.2009
Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila
Abstract
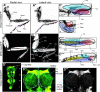
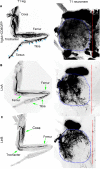
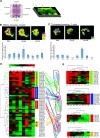
Locomotion in adult Drosophila depends on motor neurons that target a set of multifibered muscles in the appendages. Here, we describe the development of motor neurons in adult Drosophila, focusing on those that target the legs. Leg motor neurons are born from at least 11 neuroblast lineages, but two lineages generate the majority of these cells. Using genetic single-cell labeling methods, we analyze the birth order, muscle targeting, and dendritic arbors for most of the leg motor neurons. Our results reveal that each leg motor neuron is born at a characteristic time of development, from a specific lineage, and has a stereotyped dendritic architecture. Motor axons that target a particular leg segment or muscle have similar dendritic arbors but can derive from different lineages. Thus, although Drosophila uses a lineage-based method to generate leg motor neurons, individual lineages are not dedicated to generate neurons that target a single leg segment or muscle type.
Figures




References
-
- Bässler U, Büschges A. Pattern generation for stick insect walking movements: multisensory control of a locomotor program. Brain Res Brain Res Rev. 1998;27:65–88. - PubMed
-
- Bhat KM. Cell-cell signaling during neurogenesis: some answers and many questions. Int J Dev Biol. 1998;42:127–139. - PubMed
-
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech Dev. 1995;53:393–402. - PubMed
-
- Brody T, Odenwald WF. Regulation of temporal identities during Drosophila neuroblast lineage development. Curr Opin Cell Biol. 2005;17:672–675. - PubMed
-
- Burrows M. Local circuits for the control of leg movements in an insect. Trends Neurosci. 1992;15:226–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
