Database analysis of simulated and recorded electrophysiological datasets with PANDORA's toolbox
- PMID: 19475520
- PMCID: PMC2786174
- DOI: 10.1007/s12021-009-9048-z
Database analysis of simulated and recorded electrophysiological datasets with PANDORA's toolbox
Abstract
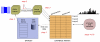
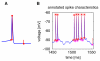
Neuronal recordings and computer simulations produce ever growing amounts of data, impeding conventional analysis methods from keeping pace. Such large datasets can be automatically analyzed by taking advantage of the well-established relational database paradigm. Raw electrophysiology data can be entered into a database by extracting its interesting characteristics (e.g., firing rate). Compared to storing the raw data directly, this database representation is several orders of magnitude higher efficient in storage space and processing time. Using two large electrophysiology recording and simulation datasets, we demonstrate that the database can be queried, transformed and analyzed. This process is relatively simple and easy to learn because it takes place entirely in Matlab, using our database analysis toolbox, PANDORA. It is capable of acquiring data from common recording and simulation platforms and exchanging data with external database engines and other analysis toolboxes, which make analysis simpler and highly interoperable. PANDORA is available to be freely used and modified because it is open-source (http://software.incf.org/software/pandora/home).
Figures
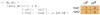
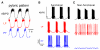




References
-
- Bean BP. The action potential in mammalian central neurons. Nature Reviews. Neuroscience. 2007;8(6):451–465. doi:10.1038/nrn2148. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

