DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition
- PMID: 19476377
- PMCID: PMC2707511
- DOI: 10.1021/cr900097c
DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition
Figures
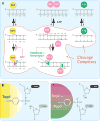
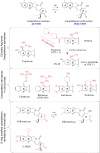
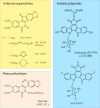
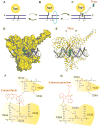


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

