Modeling inducible human tissue neoplasia identifies an extracellular matrix interaction network involved in cancer progression
- PMID: 19477427
- PMCID: PMC3050547
- DOI: 10.1016/j.ccr.2009.04.002
Modeling inducible human tissue neoplasia identifies an extracellular matrix interaction network involved in cancer progression
Abstract
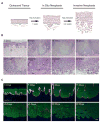
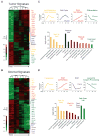
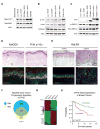
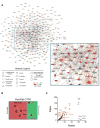
To elucidate mechanisms of cancer progression, we generated inducible human neoplasia in three-dimensionally intact epithelial tissue. Gene expression profiling of both epithelia and stroma at specific time points during tumor progression revealed sequential enrichment of genes mediating discrete biologic functions in each tissue compartment. A core cancer progression signature was distilled using the increased signaling specificity of downstream oncogene effectors and subjected to network modeling. Network topology predicted that tumor development depends on specific extracellular matrix-interacting network hubs. Blockade of one such hub, the beta1 integrin subunit, disrupted network gene expression and attenuated tumorigenesis in vivo. Thus, integrating network modeling and temporal gene expression analysis of inducible human neoplasia provides an approach to prioritize and characterize genes functioning in cancer progression.
Figures


References
-
- Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344:975–983. - PubMed
-
- Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. - PubMed
-
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. - PubMed
-
- Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van’t Veer LJ, Bartelink H, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–3743. - PMC - PubMed
-
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

