A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival
- PMID: 19477428
- PMCID: PMC2743093
- DOI: 10.1016/j.ccr.2009.03.022
A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival
Erratum in
-
A gene expression signature associated with ''K-Ras addiction'' reveals regulators of EMT and tumor cell survival.Cancer Cell. 2021 Mar 8;39(3):441-442. doi: 10.1016/j.ccell.2021.02.009. Cancer Cell. 2021. PMID: 33689705 No abstract available.
Abstract
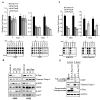
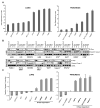
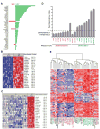
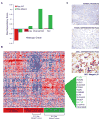
K-ras mutations occur frequently in epithelial cancers. Using short hairpin RNAs to deplete K-Ras in lung and pancreatic cancer cell lines harboring K-ras mutations, two classes were identified-lines that do or do not require K-Ras to maintain viability. Comparing these two classes of cancer cells revealed a gene expression signature in K-Ras-dependent cells, associated with a well-differentiated epithelial phenotype, which was also seen in primary tumors. Several of these genes encode pharmacologically tractable proteins, such as Syk and Ron kinases and integrin beta6, depletion of which induces epithelial-mesenchymal transformation (EMT) and apoptosis specifically in K-Ras-dependent cells. These findings indicate that epithelial differentiation and tumor cell viability are associated, and that EMT regulators in "K-Ras-addicted" cancers represent candidate therapeutic targets.
Figures



References
-
- Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, Ayala GE, Peterson LE, Ittmann M, Spencer DM. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12:559–571. - PubMed
-
- Baines AT, Lim KH, Shields JM, Lambert JM, Counter CM, Der CJ, Cox AD. Use of retrovirus expression of interfering RNA to determine the contribution of activated K-Ras and ras effector expression to human tumor cell growth. Methods Enzymol. 2006;407:556–574. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

