Genome flexibility in Neisseria meningitidis
- PMID: 19477564
- PMCID: PMC3898611
- DOI: 10.1016/j.vaccine.2009.04.064
Genome flexibility in Neisseria meningitidis
Abstract
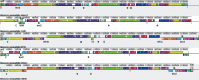
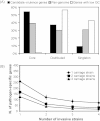
Neisseria meningitidis usually lives as a commensal bacterium in the upper airways of humans. However, occasionally some strains can also cause life-threatening diseases such as sepsis and bacterial meningitis. Comparative genomics demonstrates that only very subtle genetic differences between carriage and disease strains might be responsible for the observed virulence differences and that N. meningitidis is, evolutionarily, a very recent species. Comparative genome sequencing also revealed a panoply of genetic mechanisms underlying its enormous genomic flexibility which also might affect the virulence of particular strains. From these studies, N. meningitidis emerges as a paradigm for organisms that use genome variability as an adaptation to changing and thus challenging environments.
Figures


Similar articles
-
Living in a changing environment: insights into host adaptation in Neisseria meningitidis from comparative genomics.Int J Med Microbiol. 2007 Nov;297(7-8):601-13. doi: 10.1016/j.ijmm.2007.04.003. Epub 2007 Jun 14. Int J Med Microbiol. 2007. PMID: 17572149 Review.
-
Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis.Proc Natl Acad Sci U S A. 2008 Mar 4;105(9):3473-8. doi: 10.1073/pnas.0800151105. Epub 2008 Feb 27. Proc Natl Acad Sci U S A. 2008. PMID: 18305155 Free PMC article.
-
Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes.Microbiology (Reading). 2006 Dec;152(Pt 12):3733-3749. doi: 10.1099/mic.0.29261-0. Microbiology (Reading). 2006. PMID: 17159225
-
Metabolism and virulence in Neisseria meningitidis.Front Cell Infect Microbiol. 2014 Aug 20;4:114. doi: 10.3389/fcimb.2014.00114. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25191646 Free PMC article. Review.
-
Comparative genomics identifies the genetic islands that distinguish Neisseria meningitidis, the agent of cerebrospinal meningitis, from other Neisseria species.Infect Immun. 2002 Dec;70(12):7063-72. doi: 10.1128/IAI.70.12.7063-7072.2002. Infect Immun. 2002. PMID: 12438387 Free PMC article.
Cited by
-
Biofilm formation by the human pathogen Neisseria meningitidis.Med Microbiol Immunol. 2010 Aug;199(3):173-83. doi: 10.1007/s00430-010-0149-y. Epub 2010 Apr 8. Med Microbiol Immunol. 2010. PMID: 20376486 Review.
-
A Narrative Review of the Molecular Epidemiology and Laboratory Surveillance of Vaccine Preventable Bacterial Meningitis Agents: Streptococcus pneumoniae, Neisseria meningitidis, Haemophilus influenzae and Streptococcus agalactiae.Microorganisms. 2021 Feb 22;9(2):449. doi: 10.3390/microorganisms9020449. Microorganisms. 2021. PMID: 33671611 Free PMC article. Review.
-
Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence.J Bacteriol. 2010 Oct;192(20):5363-77. doi: 10.1128/JB.00883-10. Epub 2010 Aug 13. J Bacteriol. 2010. PMID: 20709895 Free PMC article.
-
Whole-genome sequence of the transformable Neisseria meningitidis serogroup A strain WUE2594.J Bacteriol. 2011 Apr;193(8):2064-5. doi: 10.1128/JB.00084-11. Epub 2011 Feb 4. J Bacteriol. 2011. PMID: 21296965 Free PMC article.
-
Developing insights into the mechanisms of evolution of bacterial pathogens from whole-genome sequences.Future Microbiol. 2012 Nov;7(11):1283-1296. doi: 10.2217/fmb.12.108. Future Microbiol. 2012. PMID: 23075447 Free PMC article. Review.
References
-
- Dempsey J.A., Wallace A.B., Cannon J.G. The physical map of the chromosome of a serogroup A strain of Neisseria meningitidis shows complex rearrangements relative to the chromosomes of the two mapped strains of the closely related species N. gonorrhoeae. J Bacteriol. 1995;177(22):6390–6400. - PMC - PubMed
-
- Gaher M., Einsiedler K., Crass T., Bautsch W. A physical and genetic map of Neisseria meningitidis B1940. Mol Microbiol. 1996;19(2):249–259. - PubMed
-
- Bautsch W. Comparison of the genome organization of pathogenic neisseriae. Electrophoresis. 1998;19(4):577–581. - PubMed
-
- Froholm L.O., Kolsto A.B., Berner J.M., Caugant D.A. Genomic rearrangements in Neisseria meningitidis strains of the ET-5 complex. Curr Microbiol. 2000;40(6):372–379. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

