Genome flexibility in Neisseria meningitidis
- PMID: 19477564
- PMCID: PMC3898611
- DOI: 10.1016/j.vaccine.2009.04.064
Genome flexibility in Neisseria meningitidis
Abstract
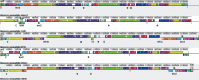
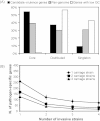
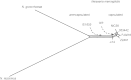
Neisseria meningitidis usually lives as a commensal bacterium in the upper airways of humans. However, occasionally some strains can also cause life-threatening diseases such as sepsis and bacterial meningitis. Comparative genomics demonstrates that only very subtle genetic differences between carriage and disease strains might be responsible for the observed virulence differences and that N. meningitidis is, evolutionarily, a very recent species. Comparative genome sequencing also revealed a panoply of genetic mechanisms underlying its enormous genomic flexibility which also might affect the virulence of particular strains. From these studies, N. meningitidis emerges as a paradigm for organisms that use genome variability as an adaptation to changing and thus challenging environments.
Figures


References
-
- Dempsey J.A., Wallace A.B., Cannon J.G. The physical map of the chromosome of a serogroup A strain of Neisseria meningitidis shows complex rearrangements relative to the chromosomes of the two mapped strains of the closely related species N. gonorrhoeae. J Bacteriol. 1995;177(22):6390–6400. - PMC - PubMed
-
- Gaher M., Einsiedler K., Crass T., Bautsch W. A physical and genetic map of Neisseria meningitidis B1940. Mol Microbiol. 1996;19(2):249–259. - PubMed
-
- Bautsch W. Comparison of the genome organization of pathogenic neisseriae. Electrophoresis. 1998;19(4):577–581. - PubMed
-
- Froholm L.O., Kolsto A.B., Berner J.M., Caugant D.A. Genomic rearrangements in Neisseria meningitidis strains of the ET-5 complex. Curr Microbiol. 2000;40(6):372–379. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

