Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion
- PMID: 19477917
- PMCID: PMC2719570
- DOI: 10.1091/mbc.e09-03-0252
Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion
Abstract
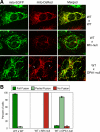
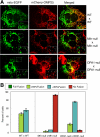
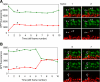
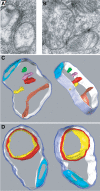
Mitochondrial fusion requires the coordinated fusion of the outer and inner membranes. Three large GTPases--OPA1 and the mitofusins Mfn1 and Mfn2--are essential for the fusion of mammalian mitochondria. OPA1 is mutated in dominant optic atrophy, a neurodegenerative disease of the optic nerve. In yeast, the OPA1 ortholog Mgm1 is required for inner membrane fusion in vitro; nevertheless, yeast lacking Mgm1 show neither outer nor inner membrane fusion in vivo, because of the tight coupling between these two processes. We find that outer membrane fusion can be readily visualized in OPA1-null mouse cells in vivo, but these events do not progress to inner membrane fusion. Similar defects are found in cells lacking prohibitins, which are required for proper OPA1 processing. In contrast, double Mfn-null cells show neither outer nor inner membrane fusion. Mitochondria in OPA1-null cells often contain multiple matrix compartments bounded together by a single outer membrane, consistent with uncoupling of outer versus inner membrane fusion. In addition, unlike mitofusins and yeast Mgm1, OPA1 is not required on adjacent mitochondria to mediate membrane fusion. These results indicate that mammalian mitofusins and OPA1 mediate distinct sequential fusion steps that are readily uncoupled, in contrast to the situation in yeast.
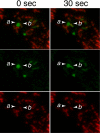
Figures


References
-
- Alavi M. V., et al. A splice site mutation in the murine Opa1 gene features pathology of autosomal dominant optic atrophy. Brain. 2007;130:1029–1042. - PubMed
-
- Alexander C., et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. - PubMed
-
- Chan D. C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006;22:79–99. - PubMed
-
- Chen H., Chomyn A., Chan D. C. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

