Transcriptional analysis of the Candida albicans cell cycle
- PMID: 19477921
- PMCID: PMC2710843
- DOI: 10.1091/mbc.e09-03-0210
Transcriptional analysis of the Candida albicans cell cycle
Abstract
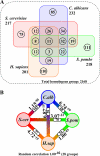
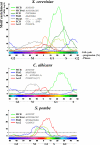
We have examined the periodic expression of genes through the cell cycle in cultures of the human pathogenic fungus Candida albicans synchronized by mating pheromone treatment. Close to 500 genes show increased expression during the G1, S, G2, or M transitions of the C. albicans cell cycle. Comparisons of these C. albicans periodic genes with those already found in the budding and fission yeasts and in human cells reveal that of 2200 groups of homologous genes, close to 600 show periodicity in at least one organism, but only 11 are periodic in all four species. Overall, the C. albicans regulatory circuit most closely resembles that of Saccharomyces cerevisiae but contains a simplified structure. Although the majority of the C. albicans periodically regulated genes have homologues in the budding yeast, 20% (100 genes), most of which peak during the G1/S or M/G1 transitions, are unique to the pathogenic yeast.
Figures




References
-
- Bachewich C., Nantel A., Whiteway M. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol. Microbiol. 2005;57:942–959. - PubMed
-
- Bähler J. Cell-cycle control of gene expression in budding and fission yeast. Annu. Rev. Genet. 2005;39:69–94. - PubMed
-
- Bennett R. J., Johnson A. D. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol. Microbiol. 2006;62:100–119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

