REPETITA: detection and discrimination of the periodicity of protein solenoid repeats by discrete Fourier transform
- PMID: 19478001
- PMCID: PMC2687986
- DOI: 10.1093/bioinformatics/btp232
REPETITA: detection and discrimination of the periodicity of protein solenoid repeats by discrete Fourier transform
Abstract
Motivation: Proteins with solenoid repeats evolve more quickly than non-repetitive ones and their periodicity may be rapidly hidden at sequence level, while still evident in structure. In order to identify these repeats, we propose here a novel method based on a metric characterizing amino-acid properties (polarity, secondary structure, molecular volume, codon diversity, electric charge) using five previously derived numerical functions.
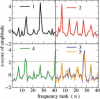
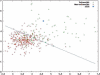
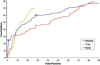
Results: The five spectra of the candidate sequences coding for structural repeats, obtained by Discrete Fourier Transform (DFT), show common features allowing determination of repeat periodicity with excellent results. Moreover it is possible to introduce a phase space parameterized by two quantities related to the Fourier spectra which allow for a clear distinction between a non-homologous set of globular proteins and proteins with solenoid repeats. The DFT method is shown to be competitive with other state of the art methods in the detection of solenoid structures, while improving its performance especially in the identification of periodicities, since it is able to recognize the actual repeat length in most cases. Moreover it highlights the relevance of local structural propensities in determining solenoid repeats.
Availability: A web tool implementing the algorithm presented in the article (REPETITA) is available with additional details on the data sets at the URL: http://protein.bio.unipd.it/repetita/.
Figures


Similar articles
-
Solenoid and non-solenoid protein recognition using stationary wavelet packet transform.Bioinformatics. 2010 Sep 15;26(18):i467-73. doi: 10.1093/bioinformatics/btq371. Bioinformatics. 2010. PMID: 20823309 Free PMC article.
-
RAPHAEL: recognition, periodicity and insertion assignment of solenoid protein structures.Bioinformatics. 2012 Dec 15;28(24):3257-64. doi: 10.1093/bioinformatics/bts550. Epub 2012 Sep 8. Bioinformatics. 2012. PMID: 22962341
-
PRIGSA: protein repeat identification by graph spectral analysis.J Bioinform Comput Biol. 2014 Dec;12(6):1442009. doi: 10.1142/S0219720014420098. J Bioinform Comput Biol. 2014. PMID: 25385083
-
TAPO: A combined method for the identification of tandem repeats in protein structures.FEBS Lett. 2015 Sep 14;589(19 Pt A):2611-9. doi: 10.1016/j.febslet.2015.08.025. Epub 2015 Aug 29. FEBS Lett. 2015. PMID: 26320412 Review.
-
State-of-the-art bioinformatics protein structure prediction tools (Review).Int J Mol Med. 2011 Sep;28(3):295-310. doi: 10.3892/ijmm.2011.705. Epub 2011 May 23. Int J Mol Med. 2011. PMID: 21617841 Review.
Cited by
-
CSpritz: accurate prediction of protein disorder segments with annotation for homology, secondary structure and linear motifs.Nucleic Acids Res. 2011 Jul;39(Web Server issue):W190-6. doi: 10.1093/nar/gkr411. Epub 2011 Jun 6. Nucleic Acids Res. 2011. PMID: 21646342 Free PMC article.
-
RepeatsDB: a database of tandem repeat protein structures.Nucleic Acids Res. 2014 Jan;42(Database issue):D352-7. doi: 10.1093/nar/gkt1175. Epub 2013 Dec 5. Nucleic Acids Res. 2014. PMID: 24311564 Free PMC article.
-
SOLeNNoID: a deep learning pipeline for solenoid residue detection in protein structures.Bioinformatics. 2025 Aug 2;41(8):btaf415. doi: 10.1093/bioinformatics/btaf415. Bioinformatics. 2025. PMID: 40689530 Free PMC article.
-
Identifying tandem Ankyrin repeats in protein structures.BMC Bioinformatics. 2014 Dec 30;15(1):6599. doi: 10.1186/s12859-014-0440-9. BMC Bioinformatics. 2014. PMID: 25547411 Free PMC article.
-
Feature-based classification of amino acid substitutions outside conserved functional protein domains.ScientificWorldJournal. 2013 Nov 17;2013:948617. doi: 10.1155/2013/948617. eCollection 2013. ScientificWorldJournal. 2013. PMID: 24348198 Free PMC article.
References
-
- Andrade MA, et al. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. - PubMed
-
- Andrade MA, et al. Homology-based method for identification of protein repeats using statistical significance estimates. J. Mol. Biol. 2000;298:521–537. - PubMed
-
- Biegert A, Soding J. De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics. 2008;24:807–814. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

