From disease ontology to disease-ontology lite: statistical methods to adapt a general-purpose ontology for the test of gene-ontology associations
- PMID: 19478018
- PMCID: PMC2687947
- DOI: 10.1093/bioinformatics/btp193
From disease ontology to disease-ontology lite: statistical methods to adapt a general-purpose ontology for the test of gene-ontology associations
Abstract
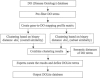
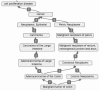
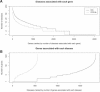
Subjective methods have been reported to adapt a general-purpose ontology for a specific application. For example, Gene Ontology (GO) Slim was created from GO to generate a highly aggregated report of the human-genome annotation. We propose statistical methods to adapt the general purpose, OBO Foundry Disease Ontology (DO) for the identification of gene-disease associations. Thus, we need a simplified definition of disease categories derived from implicated genes. On the basis of the assumption that the DO terms having similar associated genes are closely related, we group the DO terms based on the similarity of gene-to-DO mapping profiles. Two types of binary distance metrics are defined to measure the overall and subset similarity between DO terms. A compactness-scalable fuzzy clustering method is then applied to group similar DO terms. To reduce false clustering, the semantic similarities between DO terms are also used to constrain clustering results. As such, the DO terms are aggregated and the redundant DO terms are largely removed. Using these methods, we constructed a simplified vocabulary list from the DO called Disease Ontology Lite (DOLite). We demonstrated that DOLite results in more interpretable results than DO for gene-disease association tests. The resultant DOLite has been used in the Functional Disease Ontology (FunDO) Web application at http://www.projects.bioinformatics.northwestern.edu/fundo.
Figures


References
-
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Dennis G, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. - PubMed
-
- Dopazo J. Functional interpretation of microarray experiments. OMICS. 2006;10:398–410. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

